MALDI imaging — an analytical technique that detects potential biomarkers directly from tissue sections — has become popular over the last 10 years. One significant aspect is the development protocols for spatially resolved digests.
Above all, on-tissue digestion enables the analysis of peptides from formalin-fixed paraffin-embedded (FFPE) tissue — the most general type of sample in clinical pathology. Moreover, it enables the detection of larger proteins using peptides as proxies and simplifies biomarker ID using MS/MS.
Bruker established its formerly explained ImageID workflow (integrating MALDI and LC-MALDI imaging) on an innovative MALDI-TOF/TOF instrument with a 10 kHz laser. Acquisition time is reduced by a factor of 5-10, enabling the whole experiment to be completed in one working day. At this speed, ImageID serves as a robust tool for the evaluation and optimization of on-tissue digestion protocols.
| Keywords |
Instrumentation and Software |
| ImageID workflow |
rapifleX TOF/TOF |
| Protein identification in tissue |
ImagePrep |
| MALDI imaging |
TM-Sprayer |
| LC-MALDI |
ImageID software |
| FFPE tissue |
flexImaging software |
Methods
For the experiment, tissue sections (FFPE or fresh-frozen) were mounted on ITO-slides. Two serial sections were then mounted on the same slide to perform imaging and LC-MALDI experiments simultaneously on similar samples. An ImagePrep (Bruker Daltonik, Germany) or TM-Sprayer (HTX Technologies, USA) was used to apply trypsin for spatially resolved digestion. Using the same device, one section was coated with matrix after digestion and imaged on a rapifleXTM (Bruker Daltonik).
Peptides were extracted from the adjacent section with 20 µl of 0.1% TFA and isolated by nano-LC (using a 60 minute gradient split into 384 fractions). The LC-MALDI analysis was performed on a rapifleXTM in TOF/TOF configuration. LC-MALDI and imaging data were integrated on the basis of intact peptide masses after statistical recalibration of the entire spectra using the ImageID software, resulting in localized and annotated peptides (see Figure 1). Additionally, on-tissue MS/MS was used to confirm the selected peptide IDs (see Figure 3).
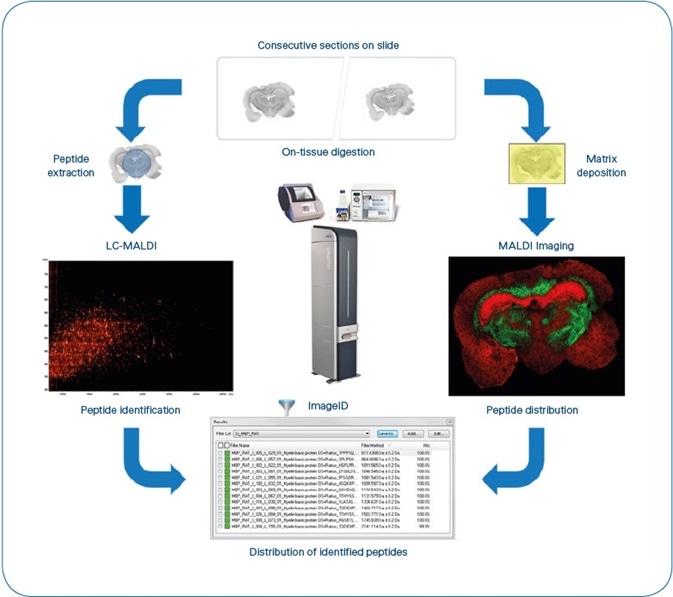
Figure 1. ImageID workflow. After matching LC-MALDI and MS Imaging data, results are available as flexImaging filter lists, grouping all peptides detected for each individual protein. Typical experiment time is 8 hours, not including antigen-retrieval for FFPE tissue. (4 hours for sample preparation, 1 hour for MALDI Imaging (Rat brain coronal section at 50 μm pixel size ~35,000 MS spectra) and 3 hours for LC-MALDI (384 fractions, ~5000 MS/MS spectra).
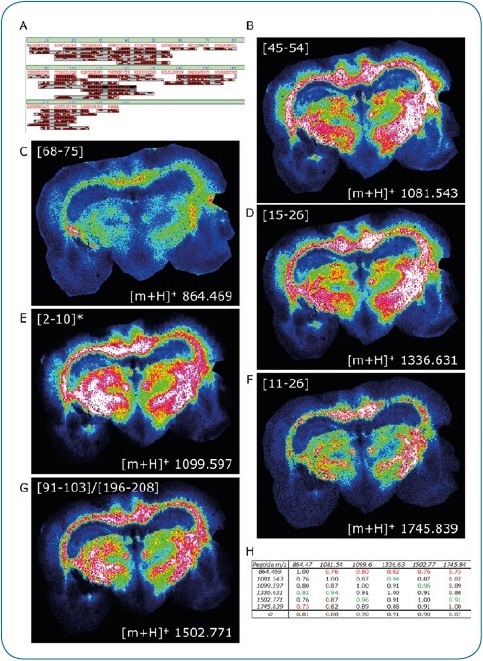
Figure 2. ImageID results. LC-MALDI analysis of peptides extracted from a coronal rat brain section achieved a 97.9% sequence coverage of Myelin basic protein, an abundant brain protein, with 42 peptides identified (A). 30 of these peptides were also matched to the corresponding MALDI image. Distribution of six peptides (B-G) is shown, which were all confirmed by on-tissue MS/MS.
For added confidence, we calculated the Pearson correlation matrix for these six images, indicating a high co-localization of all peptides (H)
* Modification at protein N-terminus determined: acetylation after methionine removal.
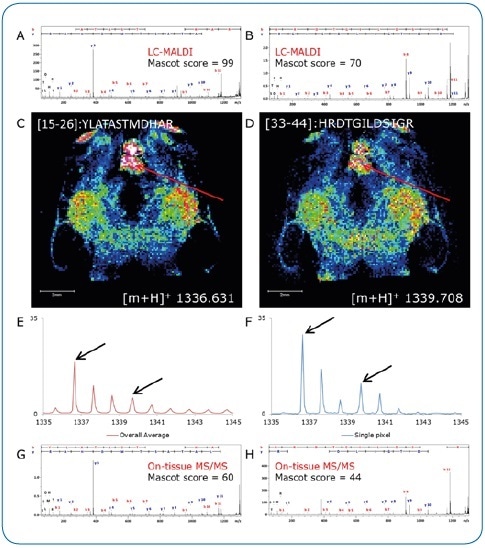
Figure 3. On-tissue MS/MS. Peptides identified by LC-MALDI (A, B) were confirmed by on-tissue measurements. A consecutive section was digested under identical conditions and peptide distributions visualized by a quick (15 minutes) imaging experiment at 100 µm pixel size (C, D). Although not clearly separated in the overall spectrum (E), two peptides (Δm/z = 3.077) could be clearly discriminated by on-tissue MS/MS at high-intensity pixels (F) using the rapifleX (G, H).
Results
Table 1. Mass accuracy on-tissue. Identifying peptides by ImageID allowed us to assess mass accuracy in MS mode
| Calculated Petide m/z |
External Calibration |
Δ m/z |
Δ ppm |
Statistical Recalibration |
Δ m/z |
Δ ppm |
| 864.469 |
864.494 |
-0.025 |
-28.9 |
864.469 |
0.000 |
0.0 |
| 1081.543 |
1081.546 |
-0.003 |
-2.8 |
1081.517 |
0.026 |
24.0 |
| 1336.631 |
1336.689 |
-0.058 |
-43.4 |
1336.656 |
-0.025 |
-18.7 |
| 1339.708 |
1339.709 |
-0.001 |
-0.7 |
1339.676 |
0.032 |
23.9 |
| 1502.771 |
1502.791 |
-0.020 |
-13.3 |
1502.754 |
0.017 |
11.3 |
| 1745.839 |
1745.854 |
-0.015 |
-8.6 |
1745.813 |
0.026 |
14.9 |
| 2141.114 |
2141.283 |
-0.169 |
-78.9 |
2141.137 |
-0.023 |
-10.7 |
| |
Ø |
-0-044 |
-25.2 |
Ø |
0.008 |
6.4 |
The rapifleX gains a solid mass accuracy of around 25 ppm with external calibration. As per Mann et al., 1993, statistical recalibration fixes major errors, thus enhancing mass accuracy to ~7 ppm on average. All calculations were executed on the average spectra of an entire imaging run.
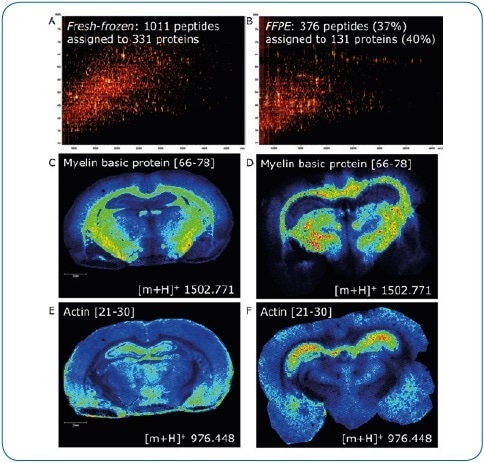
Figure 4. Fresh-frozen vs. FFPE tissue. ImageID is a powerful tool to evaluate on-tissue digests. Here, we compare fresh-frozen and FFPE rat brain tissue. Except for antigen retrieval (20 minutes at 110 °C, pH 6), digest and matrix application (using a TM-Sprayer) were identical. The lower number of peptides generated on FFPE tissue (376 or 37% compared to fresh-frozen) indicates digestion is less efficient (A, B). In addition, antigen retrieval introduces further delocalization of peptides as seen by the slight loss of lateral resolution in the FFPE experiment (D, F) when compared to fresh-frozen (C, E). Matching LC-MALDI data to the imaging data using ImageID, we could assign only 23% (235, fresh-frozen) and 57% (214, FFPE) of LC-MALDI peptides to peaks in the imaging data, indicating that extraction and incorporation of peptides during matrix application requires improvement.
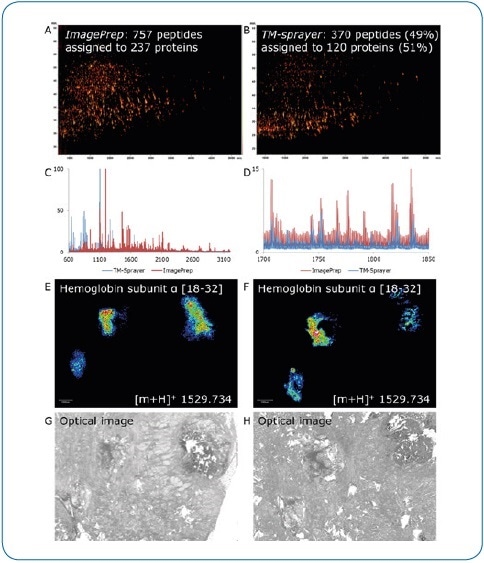
Figure 5. ImagePrep vs. TM-Sprayer. Comparison of on-tissue digestion and matrix application using serial sections of human breast cancer tissue (FFPE). Best practice protocols were used on both devices by experienced users, all MS data were acquired under identical conditions on a rapifleX. Digestion and matrix application steps appear to be less efficient using the TM-Sprayer, indicated by the lower number of peptides identified by LC-MALDI (370 peptides or 49% compared to ImagePrep) (A,B) and the lower percentage of peptides matched to the imaging run (136 or 37% vs. 399 (53%). At higher peptide masses, the ImagePrep shows more and higher intensity signals (C, D). In contrast, the TM-Sprayer provides a substantial improvement in lateral resolution. Peptide distributions (E, F) can be matched to corresponding tissue features more precisely (G, H).
Summary
Bruker has transferred the ImageID workflow to the next-generation MALDI TOF/TOF platform. Excellent mass accuracy in MS mode, integrated with LC-MALDI data and high-quality imaging enables confident matching of peptide IDs and distributions. Furthermore, the high MS/MS performance of the rapifleXTM enables direct analysis of peptides from tissue, for additional validation of ImageID matches.
In this article, ImageID was used to assess the performance of on-tissue digestion protocols, decoupling the protocol’s digestion and the matrix deposition step using the number of assigned and identified peptides respectively as useful readouts.
Conclusions
- The imageID workflow is an efficient tool for the evaluation of on-tissue digestion
- ImageID results were validated using on-tissue MS/MS
- The complete workflow, including peptide identification and tissue imaging, was completed in a single working day
- The ImageID workflow was modified to rapifleXTM
Acknowledgment
Produced from materials originally authored by Michael Becker, Anja Resemann, Janine Beckmann, Rainer Paape, Detlev Suckau Bruker Daltonik GmbH, Bremen, Germany.
Bruker would like to thank Rita Casadonte and Jörg Kriegsmann (Proteopath, Trier) for providing the breast cancer tissue samples. Bruker is grateful to Janina Oetjen (MALDI Imaging Lab, Bremen) for expert help in sample preparation. Additionally, the company would like to thank Alain Creissen and HTX for the loan of a TM-Sprayer and valuable input in protocol development
About Bruker Life Sciences Mass Spectrometry

Discover new ways to apply mass spectrometry to today’s most pressing analytical challenges. Innovations such as Trapped Ion Mobility (TIMS), smartbeam and scanning lasers for MALDI-MS Imaging that deliver true pixel fidelity, and eXtreme Resolution FTMS (XR) technology capable to reveal Isotopic Fine Structure (IFS) signatures are pushing scientific exploration to new heights. Bruker's mass spectrometry solutions enable scientists to make breakthrough discoveries and gain deeper insights.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.