Antigenic drift causes sudden changes in influenza antiviral target proteins that cloak influenza virus from the immune system of vaccinated hosts. To maintain immunological recognition against diverse influenza subtypes, a yearly formulation change for influenza vaccines and concomitant antibody treatments is required.
It is crucial to understand the binding kinetics and epitope variety of different antibodies to influenza viral antigens to cure and control outbreaks. In this article, using only 2 µL per sample, researchers perform kinetic analysis and epitope characterization of various antibodies against Influenza A nucleoprotein (NP) and hemagglutinin (HA).
Introduction
The influenza virus causes a highly infectious ailment known as “the flu.” Influenza A and B, two of the four varieties (Influenza A, B, C, and D), are the two most common flu viruses that affect humans. Both are extremely contagious and regularly cause seasonal epidemics. Influenza A viruses are also the only ones proven to cause flu outbreaks, with Influenza A viruses being linked to all five flu pandemics since 1900.
Since viruses are constantly evolving, studies of viral antibody and antigen binding and characterization are essential for managing and avoiding viral epidemics. Furthermore, these investigations contribute to our knowledge of viral antigenic drift and the variability of flu-related antibodies.
Influenza A Nucleoprotein (NP)
The influenza viral nucleoprotein is a structural protein that helps the virus replicate and adapt to its new environment. Antibodies that target NP proteins are extensively employed in ELISA, lateral flow assay (LFA), and direct fluorescent antibody tests for the identification of influenza viruses. Broad-spectrum influenza antibodies are especially needed for flu diagnostics due to the high frequency of antigenic drift or shift across various influenza strains.
Influenza A Hemagglutinin (HA)
Since antibodies can directly disrupt the virus’s attachment to the host cell receptor, the HA protein receptor-binding domain (RBD) is an important objective for vaccine development. Modifications in this protein are also necessary for vaccine development to continue.
Traditional characterization techniques
Conventional methods like ELISA and western blot (WB) involve time-consuming washing and incubation processes and rely on tags for evaluation. SPR’s information content allows for more efficient creation of viral diagnostics and treatments, as compared to standard approaches.
Without the use of tags or labels, SPR allows a complete kinetic analysis of antibody-antigen interactions to comprehend, classify, and optimize antibody therapies. It also has epitope binning capabilities, which helps researchers better comprehend epitope variety and overlap.
Kinetic analysis and epitope characterization with Alto
Alto’s digital microfluidic (DMF) powered surface plasmon resonance (SPR) technology enables researchers to examine, create, and optimize vaccines and antibody therapies, among other things. SPR’s full kinetic characterization allows for a better understanding of antibody effectiveness and antigen drift. With sample needs as low as 2 µL per protein and automated on-cartridge serial dilutions, Alto speeds up analysis.
For use in SPR-based tests, Sino Biological offers recombinant antigen preparations for all WHO-recommended vaccination strains, including HA, NA, and nucleoproteins.
This article looks at kinetic and epitope characterization tests on Alto with five distinct antibodies to two influenza A nucleoproteins (NP) and two distinct antibodies to an influenza A hemagglutinin (HA) protein, undertaken in conjunction with Sino.
Material and equipment
- Alto CBX Cartridge (KIN-CART-CBX-16)
- Alto Instrument (ALTO16)
- Nicoya Amine Coupling Kit (ALTO-R-EDCNHS)
- Running Buffer: PBS-T (0.1% Tween 20), pH 7.4 (ALTOR-PBST)
- Regeneration Buffer: 10mM Glycine-HCl pH 2.0 (ALTO-R-GLYHCl-2.0)
- Ligands and analytes: Sino Biological supplied all viral products.
Method
Direct kinetics
Alto’s on-screen setup instructions included the following steps:
- When stimulated, the Carboxyl Cartridge was loaded into the Alto instrument.
- The test was initiated by selecting the “Run Method” command.
- All reagents were pipetted into the Carboxyl Cartridge according to the on-screen guidelines.
Alto ran the commands and operations listed below automatically.
- Carboxyl sensors were primed with 10 mM Glycine-HCl for 40 seconds.
- Carboxyl surface was activated with EDC/NHS for 5 minutes.
- The Influenza A Antibody ligands were prepared in acetate pH 5 and immobilized at 20 µg/ml for 10 minutes on each of the Active Sensors.
- All sensors were blocked using an ethanolamine blocking solution for 5 minutes to quench the remaining active Carboxyl groups.
- Sensors were incubated in PBS-T for 15 minutes to collect blank injection curves.
- The experiments were executed using both single-cycle kinetics (SCK) and multi-cycle kinetics (MCK):
- Single-cycle kinetics: From a 300 nM stock concentration, Alto performed five automated 3-fold serial dilutions on the cartridge per Influenza A Protein analyte sample, yielding 1.2 nM, 3.7 nM, 11 nM, 33 nM, and 100 nM analyte samples. Commencing at the lowest concentration, influenza A Protein analyte samples were delivered in escalating concentrations with an associated duration of 180 seconds and no dissociation or regeneration between each sample. A 1200-second dissociation followed the last (highest) analyte concentration. A 60-second exposure to Glycine-HCl pH 2.0 regenerated the sensor surface.
- Multi-cycle kinetics: Alto performed five automated threefold serial dilutions on the cartridge per Influenza A Protein analyte sample from a 600 nM stock concentration, yielding 2.5 nM, 7.4 nM, 22.2 nM, 66.7 nM, and 200 nM analyte samples. The first influenza A concentration 180-second association time was used to initiate a protein analyte sample, followed by a 1200-second dissociation and a 60-second regeneration of Glycine-HCl pH 2.0. This was done for the next four concentrations.
- Completion of the test. To determine kinetic and affinity constants, binding curves were fitted to a 1:1 binding model.
Pan Influenza A Nucleoprotein Antibody binning
- Steps 1 through 9 above were followed to directly combine each one of the Pan Influenza A Nucleoprotein Antibodies to an active sensor for use as the surface antibody.
- Influenza A Protein antigen (100 nM) was presented to each capture antibody.
- Alto implemented five automated 3-fold serial dilutions on cartridge per Pan Influenza A Nucleoprotein Antibody used as the “secondary antibody” sample from the 300 nM stock concentration, producing 1.2 nM, 3.7 nM, 11 nM, 33 nM, and 100 nM analyte samples.
- The sensor surface was regenerated with a 60 seconds exposure of Glycine-HCl pH 2.0 to eliminate the secondary antibody and the antigen.
- Steps 2 through 4 were constant for each additional Pan Influenza A Nucleoprotein Antibody.
- Completion of the test. The secondary antibody was incapable to bind to the captured antigen if it contended for the same or related epitope as the surface antibody.
Results and discussion
Kinetic characterization
The binding and kinetic fits of the Influenza A Proteins to the immobilized Influenza A Antibodies resulted in good binding curves and kinetic parameter assessments. Figures 1 and 2 show a representative layer of single-cycle and multi-cycle kinetics.
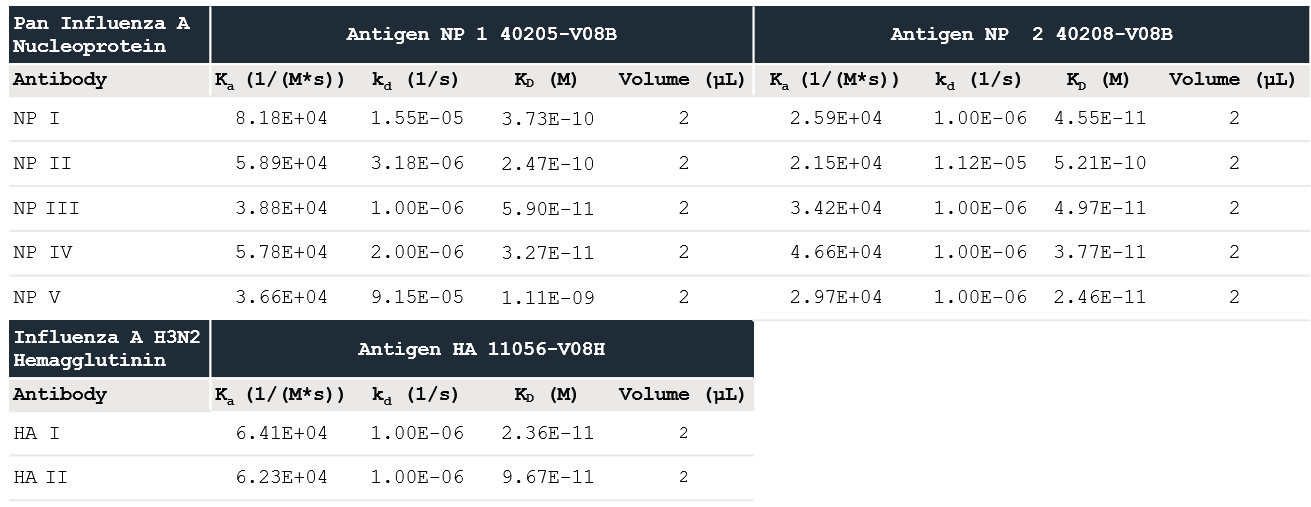
The data was fitted to a one-to-one binding model, and the determined kinetic constants are shown in Table 1. The kinetic data shows that the three Influenza A Proteins have a high affinity for all of the Influenza A Antibodies evaluated.
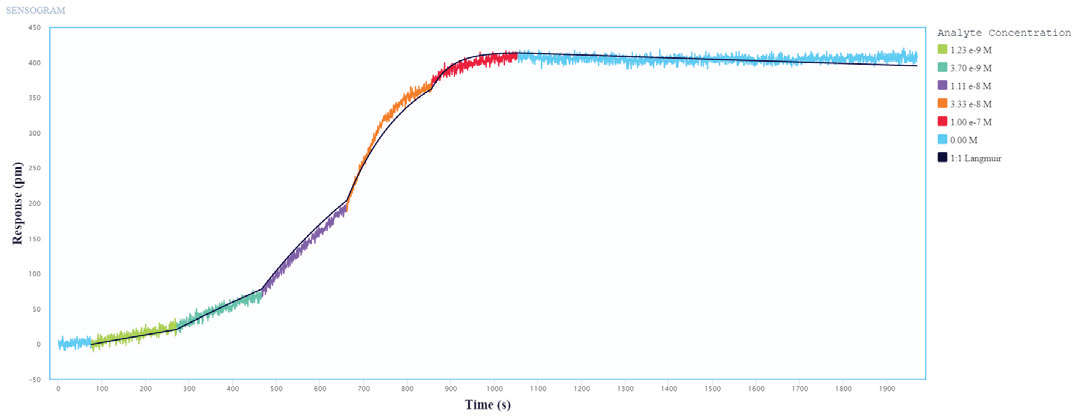
Figure 1. Single-cycle kinetics of Influenza A protein binding to immobilized Influenza A Antibodies on Alto. Influenza A protein analyte was titrated from 1.2 nM to 100 nM. The black curve is the Langmuir 1:1 binding fit model analyzed in the Nicoya Analysis Software. Image Credit: Sino Biological Inc.
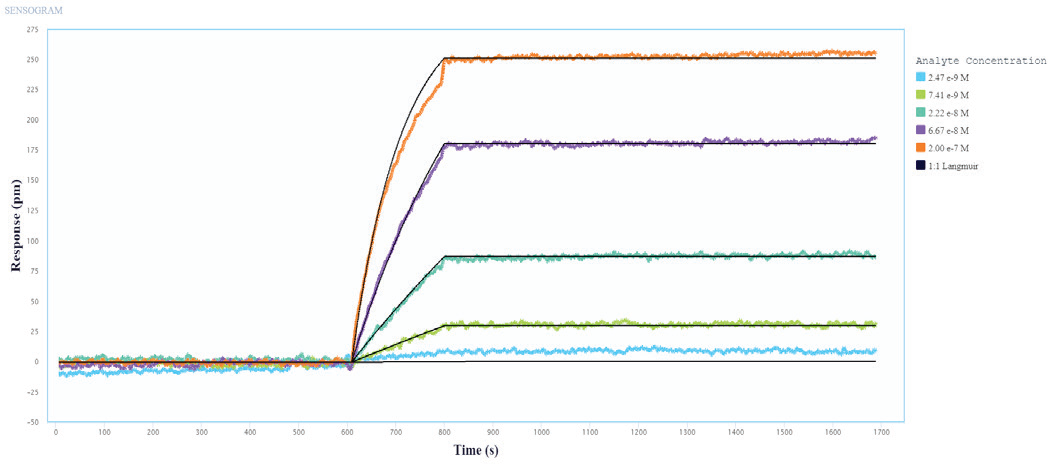
Figure 2. Multi-Cycle kinetics of Influenza A Protein binding to immobilized Influenza A Antibodies on Alto. Influenza A protein analyte was titrated from 2.4 nM to 200 nM. The black curve is the Langmuir 1:1 binding fit model analyzed in the Nicoya Analysis Software. Image Credit: Sino Biological Inc.
Table 1. Kinetic values measured using Alto data with the Nicoya analysis software. Source: Sino Biological Inc.
Epitope characterization
Sino Biological’s reagents’ epitope diversity was assessed using a sandwich assay and Capture methodology. Figures 3 and 4 show the outcomes of a 5x5 epitope bin, with “Bind” results indicating a distinct epitope focused on the antibodies. In regards to epitope characterization, kinetic values for each antibody were acquired using on-cartridge serial dilutions in the very same investigation.
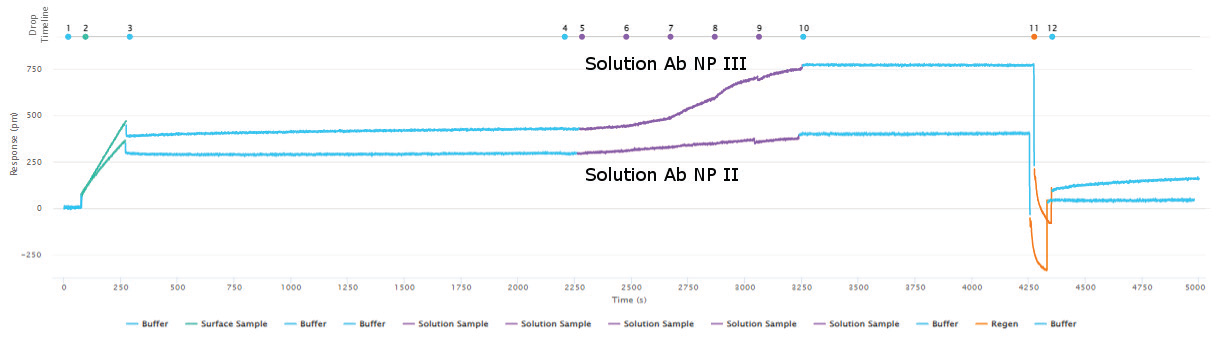
Figure 3. Epitope characterization showing capturing of the Influenza A NP Protein antigen (green curve) to the immobilized surface antibody NP I followed by binding of the solution antibody to a different epitope of the antigen (purple - NP III). A lack of binding of the solution antibody in the purple - NP II curve indicates epitope overlap of NP II with NP I. Full regeneration is achieved by 10 mM glycine HCl pH 2.0 (orange curve returning the response to the baseline before antigen capture. Image Credit: Sino Biological Inc.
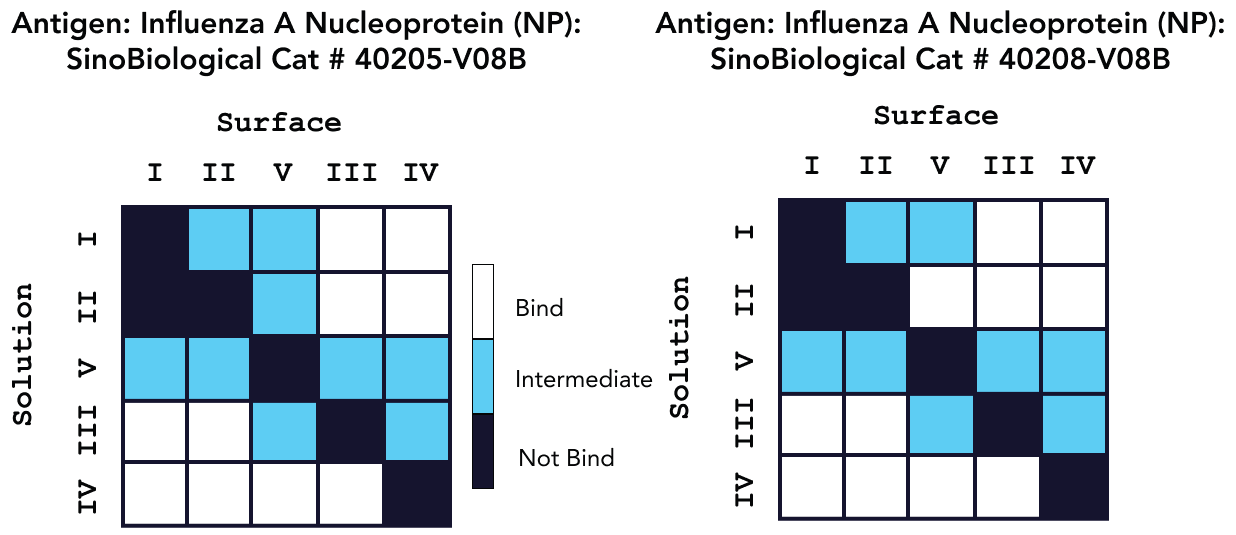
Figure 4. Epitope characterization analysis for Pan Influenza A Nucleoprotein Antibody binding to Influenza A Nucleoprotein Proteins Cat: 40205-V08B and 40208-V08B. Image Credit: Sino Biological Inc.
Conclusion
Alto was able to effectively characterize Sino Biological’s extremely specific antibodies and high-quality antigens using DMF-powered SPR to conduct kinetics and epitope analysis research. When compared to conventional characterization techniques, SPR speeds up the analysis of multiple interactions by eradicating the need for labels and lowering time-consuming preparation steps.
The information given here was collected in less than one week, highlighting the benefits of Alto’s ultra-low sample consumption combined with the use of high-quality reagents, like the amine coupling kit used in this application, and automated on-cartridge serial dilutions.
Alto’s high-throughput capabilities can speed up the research process by producing full kinetic data analysis and epitope binning experiments from as little as 2 µl of the sample while only needing less than 20 minutes of hands-on time per test.
Appendix
Table 1. Sino Biological anti-NP and anti-HA antibodies (ligands). Source: Sino Biological Inc.
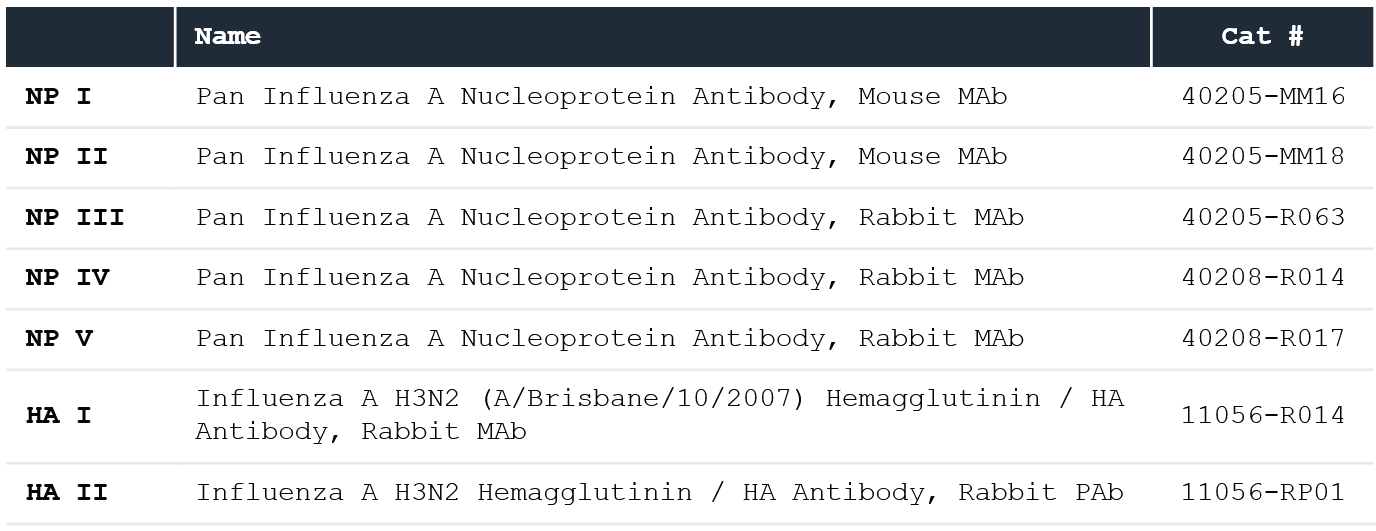
Table 2. Sino Biological recombinant proteins (analytes). Source: Sino Biological Inc.

About Sino Biological Inc.

Sino Biological is an international reagent supplier and service provider. The company specializes in recombinant protein production and antibody development. All of Sino Biological's products are independently developed and produced, including recombinant proteins, antibodies and cDNA clones. Sino Biological is the researchers' one-stop technical services shop for the advanced technology platforms they need to make advancements. In addition, Sino Biological offers pharmaceutical companies and biotechnology firms pre-clinical production technology services for hundreds of monoclonal antibody drug candidates.
Sino Biological's core business
Sino Biological is committed to providing high-quality recombinant protein and antibody reagents and to being a one-stop technical services shop for life science researchers around the world. All of our products are independently developed and produced. In addition, we offer pharmaceutical companies and biotechnology firms pre-clinical production technology services for hundreds of monoclonal antibody drug candidates. Our product quality control indicators meet rigorous requirements for clinical use samples. It takes only a few weeks for us to produce 1 to 30 grams of purified monoclonal antibody from gene sequencing.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.