The term epigenetics refers to the acquired variations in chromatin structure that occur independently of a change in the underlying sequence of DNA nucleotides. Methylation, ubiquitination, and acetylation are examples of epigenetic modifications that can change DNA accessibility to transcription machinery and consequently influence gene expression.
It has been demonstrated that the dysregulation of these epigenetic changes results in oncogenesis and cancer progression; DNA methylation, chromatin remodeling, and histone modification can regulate the cell cycle and metastasis. In contrast to genetic mutations, epigenetic alterations are said to be reversible and therefore make a potential therapeutic target.
Epigenetic Mechanism
The nucleosome is the fundamental unit of chromatin, comprising of an octamer of the histone proteins H2A, H2B, H3 and H4 (two of each) strongly linked by DNA. Chromatin structural changes caused by post-translational modifications can regulate gene expression through the formation of euchromatin or heterochromatin, which generally activate or suppress gene transcription, respectively.
Post-translational modifications include the covalent methylation (Me) and acetylation (Ac) of histone tails, and also DNA methylation. DNA methylation suppresses transcription by blocking the binding of transcription complexes to the gene promoter. The DNA from around the nucleosomes is loosened by the acetylation of histone tails, which increases the accessibility of gene promoters to transcription complexes thereby promoting transcription. Histone tail methylation can boost or suppress gene expression based on the site and extent of methylation, and also the presence of other nearby histone modifications. The transcriptional profile of genes in the vicinity is determined by the pattern of these post-translational modifications on a nucleosome. The functions of histone ubiquitination are poorly understood; however, growing evidence points to a significant role for this epigenetic modification in the DNA damage response.
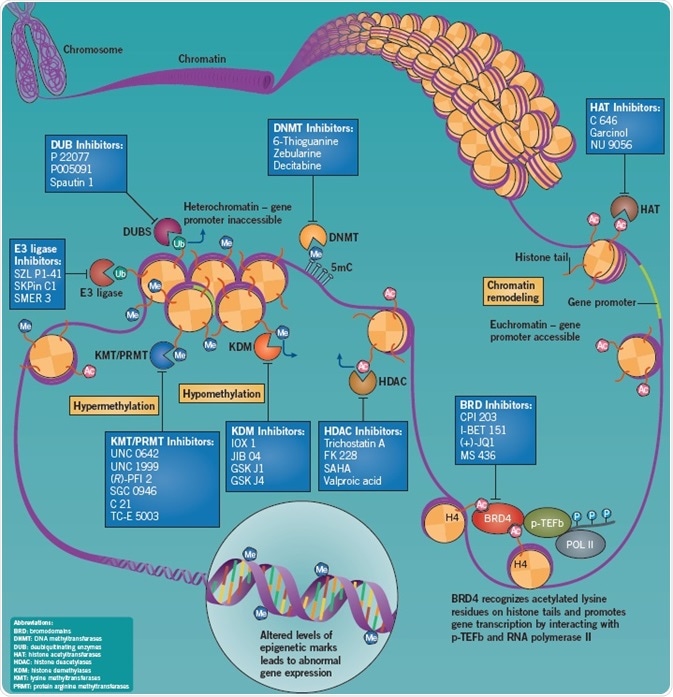
Fig 1. DNA methylation. Image credit: Tocris Bioscience
Types of Epigenetic Modifiers
Proteins that perform the epigenetic modifications described above can be considered to be either “readers”, “erasers,” or “writers”.
- Epigenetic writers catalyze the addition of epigenetic marks onto either the DNA or histone tails
- Epigenetic reader domains are effector proteins that identify and are recruited to particular epigenetic marks. Such reader domains may also be present in “eraser” and “writer” enzymes, leading to the coordination of “read-erase” or “read-write” mechanisms
- Epigenetic erasers modify gene expression by removing epigenetic marks
BRD Inhibition Suppresses Tumor Growth and Metastasis
Epigenetic “readers”, named bromodomains (BRDs), selectively identify acetylated lysine residues on histone protein tails. The BET (bromodomain and extra-terminal) bromodomain family is particularly important, and consists of the ubiquitously expressed proteins BRD2, BRD3, BRD4, and also BRDT — a testis-specific protein. BET proteins are epigenome readers that play an important role at the interface between transcriptional regulation and chromatin remodeling, and are integral in regulating the cell cycle and transcriptional elongation.
BRD4 is a crucial mediator of transcriptional elongation as it attaches to transcriptional sites of genes expressed during the M/G1 cell cycle transition and influences mitotic progression. BRD4 increases expression of genes that recruits p-TEFb to mitotic chromosomes to promote growth. In addition, it has been seen that BRD4 is considerably upregulated in both metastatic and primary melanomas. In vivo studies have demonstrated that tumor growth and metastasis is impaired by the inhibition of BRD4.
Important BRD4 inhibitors include the high affinity, potent, selective, and archetypical classic BET bromodomain inhibitor (+)-JQ1, which not only induces squamous cell differentiation but also stops tumor growth in BRD4-dependent carcinomas, including tumor growth in models of midline carcinoma cell xenograft. Recruitment of BRD3/4 to chromatin is strongly blocked by I-BET 151, which induces cell cycle arrest and apoptosis in MLL-fusion leukemia cell lines. This compound has also been demonstrated to enhance survival in rodent models of MLL-fusion leukemia.
References
- Copeland et al (2009) Protein methyltransferases as a target class for drug discovery. Nat. Rev. Drug Discov. 8 724.
- Dawson and Kouzarides (2012) Cancer epigenetics: from mechanism to therapy. Cell 150 12.
- Drouin et al (2015) Structure enabled the design of BAZ2-ICR, a chemical probe targeting the bromodomains of BAZ2A and BAZ2B. J. Med. Chem. 58 2553.
- Muller et al (2011) Bromodomains as therapeutic targets. Expert Rev. Mol. Med. 13 e29.
- Rodriguez-Paredes and Esteller (2011) Cancer epigenetics reaches mainstream oncology. Nat. Med. 17 330.
- Suva et al (2013) Epigenetic reprogramming in cancer. Science 339 1567.
- Virani et al (2012) Cancer epigenetics: a brief review. ILAR J. 253 359.
About Tocris Bioscience
Tocris Bioscience is your trusted supplier of high-performance life science reagents, including receptor agonists & antagonists, enzyme inhibitors, ion channel modulators, fluorescent probes & dyes, and compound libraries. Our catalog consists of over 4,500 research tools, covering over 400 protein targets enabling you to investigate and modulate the activity of numerous signaling pathways and physiological processes.
We have been working with scientists for over 30 years to provide the life science community with research standards, as well as novel and innovative research tools. We understand the need for researchers to trust their research reagents, which is why we are committed to supplying our customers with the highest quality products available, so you can publish with confidence.
Tocris is part of the protein sciences division of Bio-Techne, which also includes the best in class brands R&D Systems, Novus Biologicals, ProteinSimple, and Advanced Cell Diagnostics. Bio-Techne has united these brands to provide researchers with a full portfolio of research reagents, assays, and protein platforms. For more information on Bio-Techne and its brands, please visit bio-techne.com.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.