G protein-coupled receptors (GPCRs) are intrinsically allosteric proteins. Through spatially distinct, but conformationally linked, binding sites they have evolved to transduce signals from one part of the protein to another.
Endogenous activators bind to the cognate ‘orthosteric’ site of a GPCR, resulting in a conformational alteration in the GPCR which enables it to interact with intracellular transducers such as heterotrimeric G proteins. This is termed the ‘allosteric transition’. This article outlines several key insights into allosteric mechanisms of GPCR biology.
The Allosteric Nature of GPCRs
Microswitches Governing The Allosteric Transition (GPCR Activation)
The allosteric transition is controlled by a set of relatively conserved conformational contractions in the extracellular domains, regulated by conserved microswitches and water molecules. These include a smaller inward movement of TM5 and TM7 and a large outward movement of TM6, controlled by a chain of conserved intermolecular interactions or ‘microswitches’, between the transducer and orthosteric binding sites.
Key microswitches in class A GPCRs include the PIF motif, the NPxxY motif, the CWxP motif, an allosteric sodium-binding site, and the E/DRY motif. The figure illustrates these common microswitches of the class A GPCR allosteric transition, mapped onto the active-state (blue, PDB: 3PQR) and the inactive-state (white, PDB: 1F88) structures of rhodopsin.
Allosteric Modulation
The interactions between an orthosteric ligand and other conformationally linked binding sites on the same receptor are described as indirect allosteric modulation. Depending on the location of the orthosteric site, ‘Allosteric modulatory sites’ can be split into five groups. The image below uses the example M2 mAChR–iperoxo–LY2119620 structure (PDB: 4MQT).
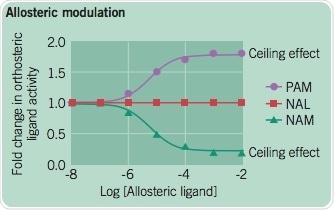
Figure 1
Impact of Oligomeric Architecture on Allosteric Signaling
GPCRs have been seen to oligomerize, adding an extra dimension to allosteric modulation, with observations outlining the potential for allosteric interactions between binding sites on GPCRs within dimeric or higher-order oligomeric formations.
In the below diagram, ovals reveal the main interfaces which have been implicated in structural biology observations, mapped onto the dimeric structure of the μOR (PDB: 4DKL).4
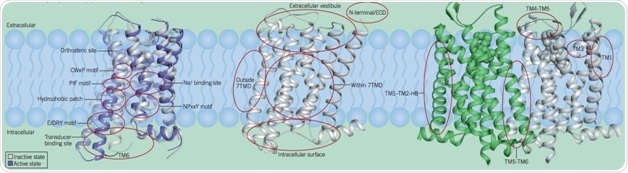
Figure 2
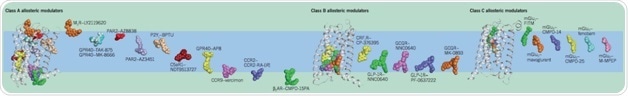
Figure 3
Diversity in the Binding Sites of Synthetic Allosteric Modulators across GPCR Classes. Chemical structures of allosteric modulators (colored spheres) mapped onto representative family members of class A (M2 mAChR, PDB: 4MQT), class B (GCGR, PDB: 5EE7) and class C (mGlu5, PDB: 4OO9) GPCRs (PDB protein database; rcsb.org).1
Conformational Mechanisms of Biased Agonism
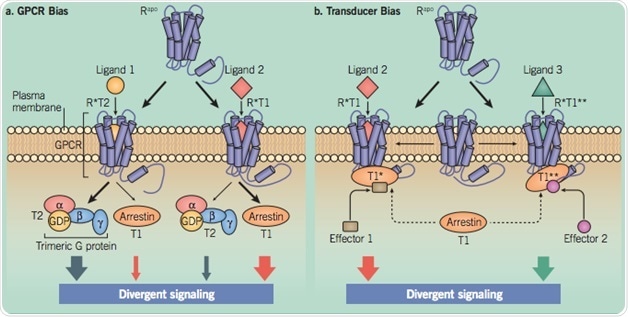
Figure 4
a. Promoting binding conditions for different transducer proteins like arrestins and G proteins, different ligands (1 and 2) begin different conformations or active states within GPCRs (R*T1 and R*T2).
b. Two different agonists (ligands 2 and 3) promote a similar active state or conformation (R*), which binds the transducer (T1), but bias emerges through allosteric communication between the transducer, receptor, and ligand, as the transducer is subject to conformational changes. Ligand 2 and 3 stabilize two different transducer conformations or active states (T1*, T1**), which leads to distinct downstream signaling events.
Key Facets of GPCR Allostery
A. Positive Allosteric Modulators (PAMs), Negative Allosteric Modulators (NAMs), and Neutral Allosteric Ligands (NALs)
There are three different modes of behavior through which allosteric modulators are characterized: PAMs NAMs or NAL2. Heightening concentrations of different allosteric modulators are titrated against a single concentration of an orthosteric ligand. A key feature of allosteric drugs is the ‘ceiling effect’; beyond this concentration of the allosteric drugs, the modulatory effect is saturated.
B. ‘Probe Dependence’
Probe dependence is another unique pharmacological characteristic of allosteric modulators whereby the direction and magnitude of the allosteric effect can alter depending on the orthosteric ligand.2
C. Receptor Subtype Selectivity
Allosteric modulators can exhibit selectivity in their capability to increase the binding of a non-selective orthosteric ligand at a certain subtype relative to other related receptor subtypes.
D. Biased Modulation
An allosteric ligand can promote multiple types of the active state. This leads to an agonist-modulator pair having different influences depending on the signaling pathway linked to each receptor conformation. The modulator enhances the potency of an agonist for pathway 1 but decreases the potency of the agonist in pathway 2 in this example.
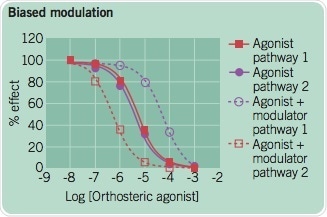
Figure 5
Therapeutic Applications of Allosteric Modulators
When compared to classic orthosteric ligands, there are many potential benefits of allosteric modulators as therapeutic drugs:
- The effect of allosteric drugs is decided by the degree of cooperativity between allosteric and orthosteric sites, which enables better control of on-target dose-related side effects. This potentially permits an allosteric medicine to be more efficacious at its GPCR target by opening up opportunities to exploit therapeutic windows which could have been too small to target with orthosteric medicines alone.
- The lower degree of amino acid conservation within allosteric sites relative to the orthosteric sites leads to greater potential for better target selectivity, with reduced off-target effects.
- Allosteric ligands may provoke endogenous agonists to have biased signaling, providing another method to target desired pathways selectively. The recent expansion in structural knowledge of GPCR allosteric sites is being included in drug-design studies.
Further to several GPCR allosteric modulators which are in clinical trials, there is growing excitement about the potential for combining allosteric drugs with existing potent, but non-selective, orthosteric drugs to enhance efficacy or to repurpose them.3,4
- ‘Pure’ NAMs or PAMs are quiescent in the absence of orthosteric endogenous agonists, exerting their effect only where and when the endogenous agonist is released. This means that where endogenous ligand levels in a given region are modified because of disease, tissue-specific conditions can be exploited.
For example, a receptor will be differentially sensitive to the allosteric drug effects to the same receptor in other tissues in the body, decreasing ‘off-tissue’ (on-target) effects in tissue with high endogenous hormone levels.
References and Further Reading
- Burger et al (2018) J. Gen. Physiol. 15 1360–1372
- Christopoulos et al (2014) Pharmacol. Rev. 66 918–947
- May et al (2007) Ann. Rev. Pharmacol. Toxicol. 47 1–51
- Thal et al (2018) Nature 559 45–53
About Tocris Bioscience
Tocris Bioscience is your trusted supplier of high-performance life science reagents, including receptor agonists & antagonists, enzyme inhibitors, ion channel modulators, fluorescent probes & dyes, and compound libraries. Our catalog consists of over 4,500 research tools, covering over 400 protein targets enabling you to investigate and modulate the activity of numerous signaling pathways and physiological processes.
We have been working with scientists for over 30 years to provide the life science community with research standards, as well as novel and innovative research tools. We understand the need for researchers to trust their research reagents, which is why we are committed to supplying our customers with the highest quality products available, so you can publish with confidence.
Tocris is part of the protein sciences division of Bio-Techne, which also includes the best in class brands R&D Systems, Novus Biologicals, ProteinSimple, and Advanced Cell Diagnostics. Bio-Techne has united these brands to provide researchers with a full portfolio of research reagents, assays, and protein platforms. For more information on Bio-Techne and its brands, please visit bio-techne.com.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.