Mitogen-activated protein kinase (MAPK) networks are essential for the transmission of extracellular signals into appropriate intracellular responses. Prototypical MAPK activation implements a three-kinase core module comprising a MAPK kinase kinase (MAPKKK or MAP3K) that phosphorylates, activating a MAPK kinase (MAP2K, MEK, or MKK).
This consequently phosphorylates, causing a dramatic increase in the activity of one or more MAPKs. Following activation, MAPKs can phosphorylate a broad range of intracellular targets, including membrane transporters, cytoskeletal elements, transcription factors, and nuclear pore proteins, in addition to further protein kinases.
Discovery of MAPKs
The MAPKs extracellular signal-regulated protein kinases 1 and 2 (ERK1/2) were originally identified as mitogen-stimulated ~42 kDa phosphoproteins during the late 1980s.1,2
Initial research indicated preparations of ERK1/2 kept the capacity to phosphorylate the model substrates myelin basic protein (MBP) and microtubule-associated protein 2 (MAP2), and reactivate phosphatase-treated ribosomal protein S6 kinase (RSK or p90).3–6
In subsequent years, discoveries revealed that the MAPK family comprised of four p38 isoforms, three c-Jun N-terminal kinases (JNKs), and ERK3 isoforms, in addition to ERK5 and ERK7 (Figure 1). The initial JNK family members were independently identified as cycloheximideactivated MBP kinases, undergoing purification for their capability of interacting with the N-terminus of the transcription factor c-Jun.7,8 p38α was identified to be a target of an inhibitor of tumor necrosis factor α (TNFα) production, an inflammatory cytokine-stimulated tyrosine phosphoprotein, and a reactivating kinase for MAPK activated protein kinase 2 (MAPKAP2 or MK2).9–11
PCR-based cloning techniques and a yeast two-hybrid screen established further JNK and p38 isoforms, in addition to ERKs 5 and 7 (reviewed by Chen et al, Sabapathy, Cuadrado & Nebreda, Nithianandarajah-Jones et al, and Drew et al; the cellular processes implicating these MAPKs are summarized in Figure 1, while comprehensive reviews by Raman et al, Dhillon et al, and Kyriakis & Avruch are also encouraged for further reading).12–19
Notably, ERK5 has recently become a focal point of therapeutic efforts20 due to its structural differences from ERK1 and 2 and its roles in cancer.21,22 While similar stimuli affect ERK1/2 and ERK5, especially growth factors, activation of ERK5 depends on divergent upstream factors.21 Regulated cross-talk and insulation are both indicators of most related MAPK pathways. Altogether, these results demonstrated that prototypical three-kinase cascades culminating in activation of a MAPK represent versatile signaling modules with the capacity to integrate a large number of inputs into an assortment of responses.
Upstream Regulation of ERK1/2
The observation that Raf and Ras mutations are widespread in several human tumors represented early evidence for ERK1/2 performing roles in proliferation and oncogenic growth.23 Research from more than one laboratory has generated an awareness that Raf isoforms are effectors of Ras small GTP binding proteins and are also upstream kinases for MAPK/ERK kinase 1 and 2 (MEK1/2), which in turn activate ERK1/2 through dual phosphorylation.24
In accordance with this initial research, an ability to drive tumorigenesis was further exhibited by the capacity of an activated mutant of MEK1 to alter cells and stimulate tumor growth in nude mice.25 Ensuing research involving dominant interfering mutants, pharmacological inhibitors of MEK1/2, and disruptions in component gene expression all demonstrated the intimate involvement of ERK1/2 in normal processes comprising cell differentiation, embryogenesis, synaptic plasticity and glucose sensing.26–31
MAP2Ks
MAP2Ks are dual-specificity kinases, able to phosphorylate both tyrosine and serine/threonine residues, which function as points of signal integration, primarily through docking site-mediated protein-protein interactions and the activity of scaffolding proteins.
Compared to MAPKs, which possess a broad array of substrates, MAP2Ks embody high-specificity; they are largely (although not exclusively) focused on phosphorylation of a small quantity of MAPK proteins.25 The model MAP2K protein, MEK1, underwent initial purification as a ~45 kDa activator of ERK1/2, while DNA-based molecular approaches subsequently enabled the identification of MEKs 2–7 (reviewed by Chen et al and Lewis et al).12,32–35
Although pharmacological inhibition of MEK1/2 is enough to virtually cease ERK1/2 signaling, a contemporary study has indicated that MEK1/2 possesses a further substrate: the proteotoxic stress response master regulator heat shock factor protein (HSF1).36 As well as identifying a link between MEK1/2 and modulation of proteomic stability, this paper also found opposing roles of ERK1/2 and MEK1/2 within the proteotoxic stress response, suggesting that ERK1/2 and MEK1/2 apply different influences on further cellular processes.
RNA sequencing experiments in mouse embryonic stem cells under conditions of pharmacological inhibition of MEK1/2 or ERK1 depletion offer moderate supporting evidence for the hypothesis that MEK embodies both independent and ERK-dependent functions, despite the fact that the continuing occurrence of ERK2.37 problematizes these experiments.
Nonetheless, it has not yet been demonstrated whether alternative MAP2Ks also possess targets outside their respective MAPKs and, in addition, how alternative substrates impact on MAPK signaling regulation.
Raf Isoforms and Other MAP3Ks
The three-kinase cascade, a module conserved from yeast to humans and which comprises a MAPK, MAP2K, and MAP3K, is the most easily recognizable property of MAPK signaling. One of the best characterized mammalian MAP3Ks is Raf, initially identified as a retroviral oncogene recognized for its contribution to cancer promotion. Three isoforms, A-Raf, B-Raf and c-Raf (or Raf-1), have been identified in mammals.38
As well as the core ~30 kDa kinase domain, Raf proteins comprise an N-terminal regulatory region of more or less identical size, which is able to bind Ras directly. Raf proteins specifically phosphorylate the MAP2Ks MEK1/2, and were originally believed to act in a tissue-specific way.
Later research, assisted by the B-Raf inhibitors which were being developed, generated the knowledge that Raf isoforms can perform dimerization to regulate signal transmission.39,40 The actions of these B-Raf inhibitors, which were not anticipated, triggered further thorough molecular analysis, which showed that dimerization can increase Raf activity and that Raf heterodimers exhibit divergent behaviors.41–43
More recent studies suggest that B-Raf dimerization is subject to feedback control by ERK1/2, which potentially explains why Raf mutants lacking the ability to dimerize maintain inappropriately elevated levels of ERK1/2 signaling.44,45 While no direct counterpart to Raf has been identified in yeast, the highly conserved nature of MAPK signaling caused researchers to search for homologs of the yeast MAP3K Ste11 among mammals.
MEKK1 was the first mammalian Ste11 homolog identified and, in spite of its ability to phosphorylate multiple MAP2Ks in vitro, much cell-based research indicates that MEKK1 primarily coordinates downstream signaling via MEKs 4 and 7 and the JNK pathway.46–51 Subsequent isolation of related cDNAs enabled the discovery of a family of enzymes (i.e. MEKKs1–4; ASK1/2; TAK1), which have since undergone review by Raman et al and Johnson et al. 18,52
As a collection, these enzymes exhibit broader substrate specificity than Raf and, it is presumed, modulate multiple MAPK pathways in a context-dependent manner. Mos and Tpl2 (Cot) represent two further enzymes that operate as MAP3Ks in the ERK1/2 pathway. These were both initially identified as proteins possessing the ability to transform cells, and both were shown to cause activation of the core cascade in specialized situations3,54.
Mos is expressed primarily in oocytes where translational recruitment of c-mos mRNA and subsequent MAPK pathway activation is essential for pronuclear formation, whereas Tpl2 undergoes stabilization in response to lipopolysaccharide.55,56 A further MAP3K family comprises homologs of the yeast protein Ste20, TAOs 1–3, which function as activators of the p38 signaling cascade.18 Figure 1 exhibits a simplified model of the organization of MAPK cascades, with emphasis made to the number of MAP2K-MAPK combinations that a specified MAP3K can modulate, while also highlighting putative nodes of cross-talk.
Properties of MAPK Cascades
A substantial amount of work on yeast MAPK cascades and the mammalian ERK1/2, p38, as well as on JNK signaling pathways, has yielded a considerable understanding of the principle properties of MAPK core modules, most notably their versatility in processing a broad array of stimuli into several responses.
Work to comprehend how three-kinase cascades accomplish such robust signaling has indicated that feedback mechanisms at more than one level underpin the evolutionary efficacy of these pathways. Furthermore, multiple biochemical mechanisms implicating pathway effectors, scaffolds, and targets serve to modulate MAPK signaling dynamics so that one core module can effectively modulate intracellular responses in a context-dependent manner (For a comprehensive review on how signaling dynamics can configure cellular responses, see Purvis & Lahav57).
One important component of ERK1/2 signaling is that two phosphorylation events on the activation loop, the first on a tyrosine residue and a second on a proximal threonine residue, are necessitated for a complete activation.
In collaboration with scaffolding proteins that bind the three-kinase cascade together, and as a result of its dual-specificity, MEK1/2 can either function as a single switch through rapid double phosphorylation or can produce a pool of monophosphorylated ERK1/2, thus multiplying the number of contexts that can ultimately attain complete pathway activation.4,58–61
The role of monophosphorylation of MAPKs might not be limited to the potentiation of a switch-like response to a graded stimulus, as research by Ashwell and colleagues has shown with p38 in T-cells.62 In this system, p38 underwent phosphorylation on the threonine activation site, but not the tyrosine, displaying altered substrate specificity, although it should be noted that this mode of activation is specific to T-cells and entails a non-canonical mechanism of p38 activation. As well as monophosphorylation, a further emergent MAPK feature is alternative phosphorylation of a threonine residue proximal to the canonical activation sites referred to above.
In 2009, the discovery was made that activated ERK2 can autophosphorylate following dimerization, causing the promotion of a distinct gene expression program in a model of cardiac hypertrophy. Subsequent biochemical characterization of a phosphomimetic mutant of the alternative phosphorylation site indicated different binding characteristics of ERK2 altered in that manner.63,64
While it is as yet unclear how divergent modes of phosphorylation help to bring about the regulation of pathway feedback, these activities likely contribute to conferring network robustness. Free ERK1/2 can enter and exit the nucleus even in the absence of pathway activation due to its interactions with nuclear pore proteins.65,66 Nonetheless, complete activation of ERK1/2 can promote dimerization and nuclear localization.67
In recent years, a small molecule, DEL 22379, capable of interrupting dimer formation even when the pathway has undergone activation, has been discovered. Notably, inhibition of dimerization seems to cause alterations to cytoplasmic ERK1/2 signaling, primarily via the disruption of interactions between ERK dimers and scaffold proteins.68,69 Further research is needed to outline how this drug impacts on the considerable nuclear activities of ERK1/2, particularly in the context of the potential role of dimerization in driving specific gene expression programs in an ERK1/2-dependent manner.63 Once doubly phosphorylated, ERK1/2 possess multiple substrates within the core-signaling pathway.
As previously explained, ERK1/2 can cause phosphorylation of Raf isoforms to block reactivation by Ras and subsequent phosphorylation of MEK1/2. This simultaneously buffers against inappropriate pathway activation and ensures a linear response to stimuli.70–75 In yeast, it has been shown that the p38 homolog Hog1p causes phosphorylation of the signaling scaffold Ste20 (homologous to mammalian PAK) progressively over time, with the authors of a contemporary study implicating Ste20 in both negative and positive feedback loops due to its capacity to function as a rheostatic effector of the MAPK pathway.76
Scaffolding Proteins
The significance of scaffold proteins has been understood for several years. However, contemporary developments to computer-aided modeling, in conjunction with biochemical work, is revealing how these proteins exert a substantial influence on cascade function as well as robustly affecting the activities and outputs of MAPK pathways.77 The yeast protein Ste5, in concert with the MAP3K Ste11, is one of the best cases of scaffold-mediated signal specificity and is needed in the activation of the downstream MAPKs Kss1 and Fus3 via Ste7, in response to pheromone.78–81
However, in response to osmotic stress, the sensor protein Sho11 allows Pbs2, the MAP2K upstream of Hog1p, to constitute a stable interaction with Ste5 and Ste11, therefore funneling distinct stimuli through the same signaling components (Figure 2).
In more recent years, a synthetic strategy was employed to “rewire” Ste5 with highly modular PDZ domains, thus improving the understanding of the impact of scaffolds on pathway dynamics.82 In accordance with previously observed evidence, this study's authors discovered that scaffolds can function as logic gates by binding core components together, in addition to modulating pathway output via the recruitment of protein phosphatases.83,84
Mammalian signaling embodies similar complexities as a result of considerable crosstalk between upstream activators, and there have been numerous indications that a singular MAP3K can modulate more than one MAPK cascade. However, because kinases themselves can function as binding hubs, tethering functions in mammalian MAPK cascades can be shared among both core pathway components as well as dedicated scaffolding proteins.
The majority of interactions with MAPKs undergo mediation by two short sequence motifs: the docking (D or DEJL) motif and the FXF (DEF) mid-1990s, with one or more of these regularly occurring in activators, scaffolds, certain phosphatases, and several substrates.85,86 JNK-interacting protein (JIP) scaffolds, which organize JNK and sometimes p38 MAPK pathways, exhibit certain functional similarities with yeast Ste5.
Slightly departing from the yeast situation, several MAP3Ks comprise docking sites that promote stable interactions with specific MAPKs. For example, MEKK1 can be bound tightly to JNKs via a docking motif.87 The scaffolding actions of kinases cannot be over-emphasized, particularly as certainly dedicated scaffolds are pseudokinases, the tethering functions of which were likely selected over enzymatic equivalents during evolution.83 The scaffold kinase suppressor of Ras (KSR) is the most commonly known instance of this. It was first identified in the sevenless eye development pathway in Drosophila and the vulval induction pathway of C. elegans.88,89
While KSR proteins are part of the Raf family, they do not possess the crucial ATP-binding lysine residue needed for functional kinase activity. Among mammals, KSR1 and KSR2 bind ERK1/2 and MEK1/2, and can allosterically activate Raf.43 Additional proteins that affect assembly and activation of MAPK pathways comprise CNK, IMP, MP-1 and Sur8.86,90–92
Activation of MAPKs from the Cell Surface
Tyrosine Kinase Receptor Activation of ERK1/2
ERK1/2 undergo activation via a broad array of stimuli that function through multiple cell surface receptors. Of these, receptor tyrosine kinase (RTK) pathways are some of the best explicated.14,18,34 Extracellular ligand-binding stimulates homo- and/or heterodimerization of RTKs, causing kinase activity to increase and producing multiple phosphotyrosine motifs on RTKs and their dimerization partners. These motifs are identified by SH2 domains on several factors, such as the adapter proteins Shc and Grb2, which also include proline-rich regions that can interact with the SH3 domain of the Ras guanine nucleotide exchange factor (GEF) son of sevenless (SOS).
Following the association with the receptor adapter protein complex, SOS triggers the exchange of GDP for GTP on Ras, promoting its interactions with a variety of downstream effectors, including Raf.
The direct interaction of Raf isoforms with Ras localizes these MAP3Ks to the plasma membrane and maintains proximity to non-receptor kinases, such as p21-activated kinases (PAKs), protein kinase C (PKC) isoforms and Src family members. In turn, these are capable of phosphorylating Ras-Raf isoforms, changing their behavior towards particular substrates, or enhancing interactions with other signaling factors.93,94 As previously mentioned, activated Raf isoforms can stimulate the MEK1/2-ERK1/2 pathway, thereby constituting the core MAPK module.
Activation of MAPKs by G-protein-coupled Receptors
Many hormones function through G-protein-coupled receptors (GPCRs) to intensify ERK1/2 activity and, just like with RTK mediated stimulation, activation transpires principally through Raf. However, in contrast to RTK signaling to ERK1/2, GPCRs generally bypass Ras and instead use a variety of mechanisms to modulate Raf activity.
Agonist binding to Gαs -coupled receptors causes adenyl cyclase to activate, and this, in turn, increases the concentration of cyclic adenosine monophosphate (cAMP). Although alterations in cAMP concentration are understood to influence ERK1/2 activity differentially, the pathway response seems to be cell-type specific or otherwise exceptionally context-dependent.95,96 A growth in cAMP stimulates protein kinase A (PKA), a signaling molecule whose activities employ extensive cross-talk with ERK1/2 signaling.97
Inhibition of ERK1/2 activity is believed to entail phosphorylation of serine 41 and serine 621 on c-Raf by PKA, which, respectively, disturbs Raf associations with Ras and 14-3-3 protein.98,99 In contrast, PKA can also increase ERK1/2 amplitude, duration, and sensitivity.97
Gαi-coupled receptors are also able to modulate ERK1/2 signaling. Early in vivo evidence suggested that Gαi -coupled receptors most likely utilize βγ subunits100,101, and indeed, contemporary research into GPCR regulation of ERK1/2 has detected an unfamiliar mode of signaling. This causes an alternative phosphorylation event which plays a causative role in cardiac hypertrophy.63
Inhibition of MAPK Pathways
MAPK Function Studied by Inhibition
A lot has been uncovered about MAPK signaling pathways via loss-of-function experiments that utilize RNA interference, small molecule inhibitors, and dominant-negative mutants. Nonetheless, because MAPKs often share common substrates and undergo activation by overlapping upstream components, the dominant-negative approach can become problematized by inhibition of multiple target pathways. Likewise, gene-silencing techniques depend to a large extent on rescue experiments due to an off-target effect, or as a means of determining whether a phenotype is pathway-dependent.
This can necessitate the targeting of more than one closely associated enzyme. In light of these factors, pharmacological inhibitors of MAPK pathway components represent both a practical alternative and a complementary tool in comprehending the functional needs of a specified pathway.102,103 Although several inhibitors bind to the ATP binding pocket common among all protein kinases, the efficacy of more specific allosteric inhibitors has driven the search for molecules that bind to different pockets on kinase surfaces.104 MAPK pathway inhibitors that block essential protein-protein interactions are undergoing development. This reflects enhanced knowledge regarding how kinase signaling networks operate.
ERK1/2 Pathway Inhibitors
Due to the high degree of feedback control demonstrated by the Raf-MEK-ERK pathway, the direct inhibition of ERK1/2 represents an appealing strategy that can be utilized concurrently with additional inhibitors of upstream factors. The small molecule FR 180204 competes for the ATP-binding pocket of ERK1/2, with IC50 values of 0.31 and 0.14 μM, respectively. However, ascertaining its practical utility is complex as a result of its targeting strategy.105
Contrastingly, the allosteric inhibitor SCH772984 seems to cause exceptional inhibition of the activity of ERK1/2 (with IC50 values of 0.004 and 0.001 μM, respectively) while also preventing reactivation by MEK1/2. This is possible via the induction of a conformational alteration which renders ERK1/2 refractory to feedback-regulated stimulation.106 The inhibitor DEL-22379, which targets and interrupts ERK dimerization rather than its catalytic activity, is somewhat related to this.69
The MEK1/2 inhibitors PD 98059 and U0126 became available in the mid-1990s, enabling researchers to interfere with the ERK1/2 pathway effectively.107 PD 98059 was identified via an in vitro kinase activation assay, while U0126 was discovered in a cell-based assay as an inhibitor of AP-1 transcriptional activity.104,108,109
Both drugs inhibit activation of MEK1/2 and are of utility at low micromolar concentrations, but necessitate higher concentrations to inhibit already activated MEK1/2 in cells.103 U0126 and PD 98059 bind externally to the ATP binding site of MEK1/2 and select for a low activity conformation, thus shifting and maintaining the protein in an inactive state. As a result of their binding mode, these drugs represent some of the most selective inhibitors around.
The only other kinase affected is the associated MAP2K MEK5, which is blocked at only slightly higher concentrations than MEK1/2. Following this, several MEK1/2 inhibitors, including ARRY-142886 (AZD6244), PD 198306, and PD 0325901 have undergone development. These embody increased potency and exhibit variable selectivity towards MEK5.110–113
PD 0325901 blocks MEK1/2 activation of ERK1/2 in cells at concentrations as low as 25 nM, but is unable to block a large panel of other protein kinases at over one hundred times the concentration.103
Nonetheless, consistent with a ~10-fold greater potency towards MEK1/2 than MEK5, ERK5 activation is blocked at low micromolar concentrations by PD 035901 (for more on ERK5 inhibition, see Chen et al).114 The efficacy of allosteric inhibitors of MEK1/2 has gone beyond the laboratory, with a number of MEK1/2 targeting compounds undergoing contemporary development as therapeutics.110,113,115–119
A further expanding category of compounds that can inhibit the ERK1/2 pathway is being directed against Raf isoforms. In particular, B-Raf has come to the fore as an interesting target as a result of the high prevalence of B-Raf V600E mutations in melanomas.
A number compounds have been identified that primarily interact with this mutant form, such as dabrafenib, encorafenib, vemurafenib, GDC-0879 and PLX-4720.120–124 Further drugs that target Raf embody lower selectivity, with compounds such as AZ 628, RO5126766 and Sorafenib blocking more than one kinase.125–127 Likewise, GW5074, LY3009120, RAF265 (CHIR-265), SB 590885, TAK-632 and ZM 336372 mainly affect multiple isoforms of Raf.118,128–133
JNK Pathway Inhibitors
SP 600125 is the most commonly utilized inhibitor of the JNK signaling pathway. Although it inhibits JNKs at concentrations between 50–100 nM, this compound also blocks several additional protein kinases with more or less equal potency.134 CEP 1347 (KT-7515) inhibits the JNK pathway, but it specifically inhibits the upstream mixed lineage kinases (MLKs), and therefore does not inhibit JNKs which are modulated in an MLK-independent manner.135
p38 pathway inhibitors
Several p38 inhibitors, such as SB 203580, have undergone development utilizing pyridinyl imidazoles as lead compounds.136,137 Crystallographic research indicates that SB 203580 obstructs ATP binding via interaction with the active site of p38, although this is only for the α and β isoforms and not p38 γ and δ.138 An important aspect of the ATP binding pocket affecting inhibitor selectivity is the gatekeeper residue, which is a threonine residue at position 106 for p38 α and β.139
The smaller size of the gatekeeper (in contrast to the glutamine and methionine side chains which occur in p38 γ and δ) permits accommodation of larger compounds in the active site.140 As a result of their small gatekeeper side chains, Raf isoforms can also interact with certain p38 inhibitors.141
BIRB 796, a diaryl urea compound that is not structurally related to SB 203580, blocks all four p38 isoforms by indirectly competing with the binding of ATP.142 There are several p38 pathway inhibitors undergoing development as therapeutics, such as pexmetinib (ARRY 614), losmapimod and ralimetinib (LY 2228820).143–149
Future Prospects
The last few years have witnessed an enhanced understanding that protein kinases represent more than phosphorylating enzymes; they are multifunctional molecules developed to process key biological information. Considering the significant roles of MAPK pathways in both normal signal transmission and a multitude of disease states, the examination of these core modules will continue to increase fundamental knowledge of cellular signaling. MAPK networks depend on multiple kinases operating in a variety of ways, producing feedback loops, switches, oscillators, and redundancies, and harnessing the activities of non-kinase proteins to contribute to robust signaling.
The evolutionary efficacy of these pathways is based on their versatility and adaptability, and although a lot has been discovered about the roles of these pathways in certain biological operations, contemporary studies continue to focus on the spatio-temporal dynamics underpinning context-dependent signaling.
As mass spectrometry, microscopy and deep sequencing techniques undergo continuing improvements, the scale of focus will carry on shifting away from individual pathways and towards entire signaling networks. In particular, pharmacological manipulation will continue to represent an essential tool for the assessment of the functionality of MAPK signaling, thus enabling enhanced therapeutic strategies that target numerous features of these multifaceted networks.
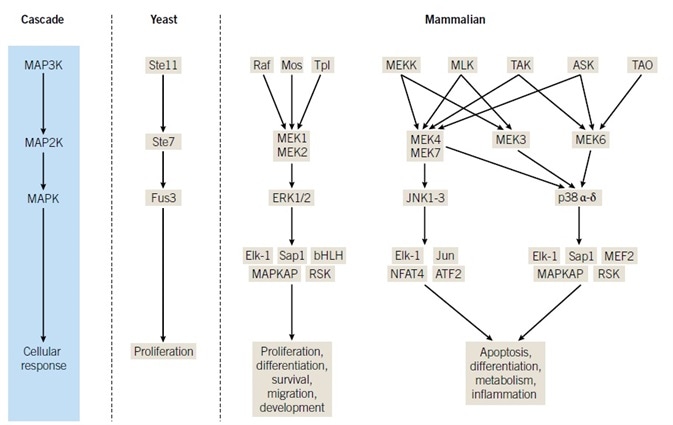
Figure 1. Regulation by Scaffolding
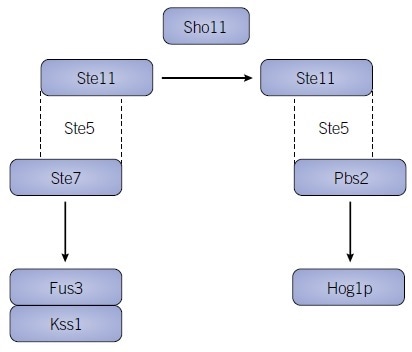
Figure 2. Regulation by Scaffolding
References and Further Reading
- Ray and Sturgill (1987) Proc.Natl.Acad.Sci. USA 84 1502.
- Sturgill et al. (1988) Nature 334 715.
- Ahn and Krebs (1990) J.Biol.Chem. 265 11495.
- Boulton et al. (1991) Biochemistry 30 278.
- Boulton et al. (1990) Science 249 64.
- Gotoh et al. (1990) Eur.J.Biochem. 193 661.
- Hibi et al. (1993) Genes.Dev. 7 2135.
- Kyriakis and Avruch (1990) J.Biol.Chem. 265 17355.
- Han et al. (1994) Science 265 808.
- Lee et al. (1994) Nature 372 739.
- Rouse et al. (1994) Cell 78 1027.
- Chen (2001) Chem.Rev. 101 2449–2476.
- Cuadrado and Nebreda (2010) Biochem.J. 429 403.
- Dhillon et al. (2007) Oncogene 26 3279.
- Drew et al. (2012) Biochim.Biophys.Acta. 1825 37.
- Kyriakis and Avruch (2012) Physiol.Rev. 92 689.
- Nithianandarajah-Jones et al. (2012) Cell Signal. 24 2187.
- Raman et al. (2007) Oncogene 26 3100.
- Sabapathy (2012) Prog.Mol.Biol.Transl.Sci. 106 145.
- Myers et al. (2016) ACS.Comb.Sci. 18 444.
- Nishimoto and Nishida (2006) EMBO Rep 7 782.
- Nithianandarajah-Jones (2014) Biochem.Soc.Trans. 42 1584.
- Land et al. (1983) Science 222 771.
- Leevers and Marshall (1992) EMBO J 11 569.
- Mansour et al. (1994) Science 265 966.
- English and Sweatt (1996) J.Biol.Chem. 271 24329.
- Khoo and Cobb (1997) Proc.Natl.Acad.Sci. USA 94 5599.
- Kunath et al. (2007) Development 134 2895.
- Lawrence et al. (2008) Proc.Natl.Acad.Sci. USA 105 13315.
- Samuels et al. (2008) J.Neurosci. 28 6983.
- Traverse et al. (1992) Biochem.J. 288 (Pt 2), 351.
- Ahn et al. (1991) J.Biol.Chem. 266 4220.
- Crews and Erikson (1992) Proc.Natl.Acad.Sci. USA 89 8205.
- Lewis et al. (1998) Adv.Cancer.Res. 74 49.
- Seger et al. (1992) J.Biol.Chem. 267 14373.
- Tang et al. (2015) Cell 160 729
- Liu et al. (2016) Genom.Data 7 73.
- Sutrave et al. (1984) Nature 309 85.
- Garnett et al. (2005) Mol.Cell 20 963.
- Weber et al. (2001) Cancer Res. 61 3595.
- Karreth et al. (2009) Mol.Cell 36 477.
- Poulikakos et al. (2010) Nature 464 427.
- Rajakulendran et al. (2009) Nature 461 542.
- Joseph et al. (2010) Proc.Natl.Acad.Sci. USA 107 14903.
- Lito et al. (2012) Cancer Cell 22 668.
- Lange-Carter et al. (1993) Science 260 315.
- Minden et al. (1994) Science 266 1719.
- Xia et al. (2000) Proc.Natl.Acad.Sci. USA 97 5243.
- Xu et al. (1997) Mol.Endocrinol. 11 1618–.
- Xu et al. (1996) Proc.Natl.Acad.Sci. USA 93 5291.
- Yujiri et al. (1998) Science 282 1911.
- Johnson et al. (2005) Curr.Opin.Chem.Biol. 9 325.
- Miyoshi et al. (1991) Mol.Cell.Biol. 11 4088.
- Papkoff et al. (1982) Cell 29 417.
- Beinke et al. (2004) Mol.Cell.Biol. 24 9658.
- Mendez et al. (2000) Nature 404 302.
- Purvis et al. (2013) Cell 152 945.
- Huang and Ferrell (1996) Proc.Natl.Acad.Sci. USA 93 10078.
- Iwamoto et al. (2016) Sci.Signal 9 ra13.
- Robbins et al. (1993) J.Biol.Chem. 268 5097.
- Robbins and Cobb (1992) Mol.Biol.Cell 3 299.
- Mittelstadt et al. (2009) J.Biol.Chem. 284 15469.
- Lorenz et al. (2009) Nat.Med. 15 75.
- McReynolds et al. (2016) Biochemistry 55 1909.
- Kosako et al. (2009) Nat.Struct.Mol.Biol. 16 1026.
- Whitehurst et al. (2002) Proc.Natl.Acad.Sci. USA 99 7496.
- Khokhlatchev et al. (1998) Cell 93 605.
- Casar et al. (2008) Mol.Cell 31 708.
- Herrero et al. (2015) Cancer.Cell 28 170.
- Mansour et al. (1994) J.Biochem. 116 304.
- Morrison and Davis (2003) Annu.Rev.CellDev. Biol 19 91.
- Ritt et al. (2010) Mol.Cell.Biol. 30 806.
- Fritsche-Guenther et al. (2011) Mol.Syst.Biol. 7 489.
- Sturm et al. (2010) Sci.Signal. 3 ra90.
- Duan and Cobb (2010) Proc.Natl.Acad.Sci. USA 107 22314.
- English et al. (2015) Sci.Signal. 8 ra5.
- Witzel et al. (2012) Front.Physiol. 3 475.
- Frost et al. (1997) EMBO J 16 6426–6438.
- O’Rourke and Herskowitz (1998) Genes.Dev. 12 2874.
- Posas and Saito (1997) Science 276 1702.
- Bhattacharyya et al. (2006) Science 311 822.
- Ryu and Park (2015) Sci.Signal. 8 ra66.
- Coyle et al. (2013) Cell 154 875.
- Park et al. (2003) Science 299 1061.
- Tanoue et al. (2000) Nat.Cell Biol. 2 110.
- Therrien et al. (1999) Proc.Natl.Acad.Sci. USA 96 13259.
- Xu and Cobb (1997) J.Biol.Chem. 272 32056.
- Sundaram and Han (1995) Cell 83 889.
- Therrien et al. (1995) Cell 83 879.
- Li et al. (2000) Genes.Dev. 14 895.
- Matheny et al. (2004) Nature 427 256.
- Schaeffer et al. (1998) Science 281 1668.
- Hamilton et al. (2001) J.Biol.Chem. 276 29079.
- King et al. (1998) Nature 396 180.
- English et al. (1999) Exp.Cell Res. 253 255.
- Gerits et al. (2008) Cell Signal. 20 1592.
- Stefan et al. (2011) Nat.Commun. 2 598.
- Kikuch and Williams (1996) J.Biol.Chem. 2
- Yip–Schneider et al. (2000) Biochem.J. 351 151.
- Koch et al. (1994) J.Biol.Chem. 269 6193.
- Luttrell et al. (1996) J.Biol.Chem. 271 19443.
- Jensen et al. (2016) CellSyst. 2 112.
- Bain et al. (2007) Biochem.J. 408 297.
- Favata et al. (1998) J.Biol.Chem. 273 18623.
- Ohori, M et al. (2005) Biochem.Biophys.Res.Commun. 336 357.
- Morris et al. (2013) Cancer Discov. 3 742.
- Wauson et al. (2013) Biochemistry 52 5164.
- Alessi et al. (1995) J.Biol.Chem. 270 27489.
- Dudley et al. (1995) Proc.Natl.Acad.Sci. USA 92 7686.
- Barrett et al. (2008) Bioorg.Med.Chem.Lett. 18 6501.
- Ciruela et al. (2003) Br.J.Pharmacol. 138 751.
- Davies et al. (2007) Mol.Cancer Ther. 6 2209.
- Yeh et al. (2007) Clin.Cancer Res. 13 1576.
- Chen et al. (2016) Acta.Crystallogr.D.Struct.Biol. 72 682.
- Adjei et al. (2015) Clin.Cancer Res. 22 2368.
- Do et al. (2015) Invest. New Drugs 33 720.
- Hatzivassiliou et al. (2013) Nature 501 232.
- Tolcher et al. (2015) Cancer Chemother.Pharmacol. 75 183.
- Watanabe et al. (2016) Cancer Chemother.Pharmacol. 77 1157.
- Bollag et al. (2012) Nat.Rev.Drug Discov. 11 873.
- Hoeflich et al. (2009) Cancer Res. 69 3042.
- Li et al. (2016) Cancer Lett. 370 332.
- Rheault et al. (2013) ACS.Med.Chem.Lett. 4 358.
- Riechardt et al. (2015) Br.J.Ophthalmol. 99 1739.
- Wada et al. (2014) PLoS One 9 e113217.
- Wilhelm et al. (2006) Nat.Rev.Drug Discov. 5 835.
- Adnane et al. (2006) Methods Enzymol. 407 597.
- Barollo et al. (2014) Invest.New Drugs 32 626.
- Chin et al. (2004) J.Neurochem. 90 595.
- Garcia–Gomez et al. (2013) Invest.New Drugs 31 200.
- Henry et al. (2015) J.Med.Chem. 58 4165.
- Kunnimalaiyaan et al. (2007) Surgery 142 959–964; discussion 959
- Okaniwa et al. (2013) J.Med.Chem. 56 6478.
- Bennett et al. (2001) Proc.Natl.Acad.Sci. USA 98 13681.
- Maroney et al. (2001) J.Biol.Chem. 276 25302.
- Cuenda et al. (1995) FEBS.Lett. 364 229.
- Tong et al. (1997) Nat.Struct.Biol. 4 311.
- Lisnock et al. (1999) Biochemistry 38, 3456.
- Eyers et al. (1998) Chem.Biol. 5 321.
- Gum et al. (1998) J.Biol.Chem. 273 15605.
- Hall-Jackson et al. (1999) Oncogene 18 2047.
- Kuma et al. (2005) J.Biol.Chem. 280 19472.
- Tate et al. (2013) J.Biol.Chem. 288 6743.
- Damjanov et al. (2009) Arthritis.Rheum. 60 1232.
- Garcia-Manero et al. (2015) Clin.Cancer Res. 21 985. .
- Selness et al. (2011) Bioorg.Med.Chem.Lett. 21 4066.
- Triantaphyllopoulos et al. (2010) Clin.Exp.Rheumatol. 28 176.
- Azevedo et al. (2012) Acta.Crystallogr.D.Biol.Crystallogr. 68 1041.
- Miwatashi et al. (2005) J.Med.Chem. 48 5966
About Tocris Bioscience
Tocris Bioscience is your trusted supplier of high-performance life science reagents, including receptor agonists & antagonists, enzyme inhibitors, ion channel modulators, fluorescent probes & dyes, and compound libraries. Our catalog consists of over 4,500 research tools, covering over 400 protein targets enabling you to investigate and modulate the activity of numerous signaling pathways and physiological processes.
We have been working with scientists for over 30 years to provide the life science community with research standards, as well as novel and innovative research tools. We understand the need for researchers to trust their research reagents, which is why we are committed to supplying our customers with the highest quality products available, so you can publish with confidence.
Tocris is part of the protein sciences division of Bio-Techne, which also includes the best in class brands R&D Systems, Novus Biologicals, ProteinSimple, and Advanced Cell Diagnostics. Bio-Techne has united these brands to provide researchers with a full portfolio of research reagents, assays, and protein platforms. For more information on Bio-Techne and its brands, please visit bio-techne.com.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.