Sponsored Content by SartoriusReviewed by Maria OsipovaFeb 19 2026
Protein-Protein Interactions (PPIs) are critical for biological systems to function properly. Strong interactions maintain structural integrity and function, whereas weaker interactions enable flexibility, allowing cells to adapt to changing conditions.
Understanding PPIs is critical for advancing research in cell biology and medication development.
Protein-protein interaction assays
Protein-protein interaction assays are roughly categorized into three major categories:
- In Vitro - This approach takes place outside the cell in a controlled laboratory environment. The techniques used are affinity chromatography, protein fragment complementation, X-ray crystallography, NMR, phage display, spectroscopy, protein assays, and biomolecular interaction studies.
- In Vivo - These approaches observe and analyze biological processes in living cells or organisms.
- In Silico – These approaches involve computer simulations and computational models. They include sequence- and structure-based approaches, in-silico two-hybrid procedures, gene expression-based methods, chromosome proximity and gene fusion techniques, phylogenetic profiling, and tree-building tools.
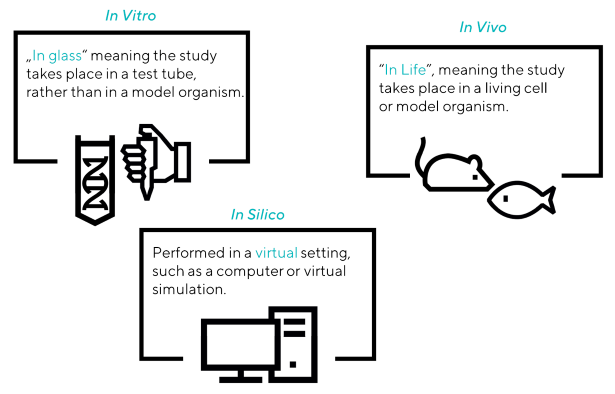
Image Credit: Sartorius
Types of protein-protein interactions
PPIs are characterized according to stability, longevity, and the types of proteins involved. Protein-protein interactions are classified in numerous ways:
They can be classified as obligatory (in which one or more proteins are unstable in vivo unless they interact and form a certain protein complex) or non-obligate interactions (in which the proteins can exist separately).
Non-obligate interactions are classified as permanent or transitory based on the stability of the complex they generate, while transient interactions can be weak or strong.
Because most obligate interactions are permanent and most non-obligate interactions are ephemeral, the terms obligate and permanent are frequently used interchangeably in literature.
Transient PPI regulates the bulk of cellular functions. As a result, much of the PPI research focuses on this form of interaction.
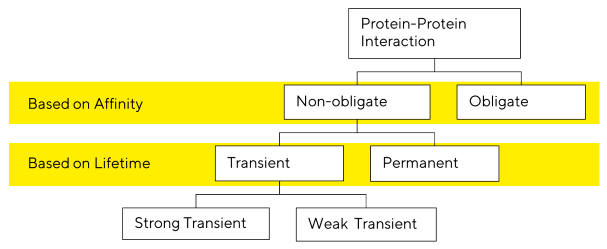
Image Credit: Sartorius
Signal transmission and metabolism are dynamic processes that rely on weaker, transitory interactions regulated by conditions such as phosphorylation or conformational changes. These weak interactions contribute to the formation of multi-enzyme complexes and to the shaping of cellular morphology and motility.
Strong PPIs, on the other hand, result in stable, multi-subunit complexes with dissociation constants that are typically nanomolar or less. RNA polymerase is a well-studied complex with significant interactions among its subunits.
Another example is the interaction of the anti-apoptotic protein Bcl-2 with the pro-apoptotic protein Bax, which regulates cell survival and death. Disruption of this balance is a major determinant in cancer development.
Protein-protein interactions as drug targets
Many medications target PPIs to disrupt or increase certain interactions, resulting in therapeutic benefits in diseases such as cancer. Small molecules, peptides, and monoclonal antibodies (mAbs) are examples of interaction modulators developed using various screening and optimization strategies.
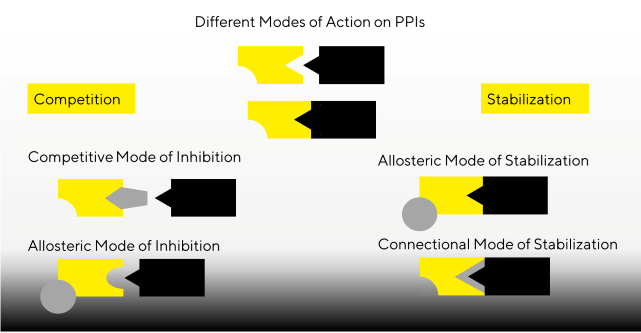
Image Credit: Sartorius
Monoclonal antibodies targeting the PD-1/PD-L1 pathway, for example, have transformed cancer immunotherapy by inhibiting this immune checkpoint and increasing antitumor responses.
Similarly, Nutlin-3 inhibits the interaction between MDM2 and p53, limiting p53 degradation and allowing its tumor-suppressive actions to take effect. This small molecule is now undergoing clinical trials and shows potential in cancer treatment.
Methods for analyzing protein-protein interactions
PPIs can be analyzed using both endpoint and real-time approaches. End-point assays offer information about a single time point and are useful for detecting interactions. Real-time approaches, on the other hand, provide the most comprehensive picture of the PPI by capturing dynamic interactions and detailed kinetics.
- Affinity-based approaches, such as co-immunoprecipitation (co-IP) and pull-down experiments, separate proteins and their binding partners from complicated mixtures like cell lysates. The enzyme-linked immunosorbent assay (ELISA) is a widely used labeled endpoint assay that measures the steady-state binding affinity of purified proteins but does not capture the dynamics of the interaction, i.e., the kinetics of how quickly the interaction formed or dissociated, as provided by on- and off-rates.
- The yeast two-hybrid genetic approach detects PPIs by triggering a transcription factor when two proteins interact.
- Real-time, label-free biophysical methods like surface plasmon resonance (SPR), biolayer interferometry (BLI), and isothermal titration calorimetry (ITC) can provide insights into binding rates and affinities, allowing researchers to optimize therapeutic targeting. These approaches are also label-free, which simplifies processes and eliminates the risk of labeling interference.
The method chosen is determined by several parameters, including target size, sample use, and throughput requirements. It is also critical to examine the interaction type, as weak or temporary interactions may be lost in some protocols.
In some cases, an ELISA provides sufficient yes/no information about the interaction of interest. However, for a more in-depth understanding of dynamic interactions, label-free, real-time data is frequently required.
References
- Acuner Ozbabacan, S.E., et al. (2011). Transient protein-protein interactions. Protein Engineering Design and Selection, 24(9), pp.635–648. DOI: 10.1093/protein/gzr025. https://academic.oup.com/peds/article-abstract/24/9/635/1556325?redirectedFrom=fulltext.
- Berggård, T., Linse, S. and James, P. (2007). Methods for the detection and analysis of protein–protein interactions. PROTEOMICS, 7(16), pp.2833–2842. DOI: 10.1002/pmic.200700131. https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/pmic.200700131.
- Goncearenco, A., et al. (2017). Exploring Protein-Protein Interactions as Drug Targets for Anti-cancer Therapy with In Silico Workflows. Methods in Molecular Biology, pp.221–236. DOI: 10.1007/978-1-4939-7201-2_15. https://link.springer.com/protocol/10.1007/978-1-4939-7201-2_15.
- Lu, H., et al. (2020). Recent advances in the development of protein–protein interactions modulators: mechanisms and clinical trials. Signal Transduction and Targeted Therapy, 5(1). DOI: 10.1038/s41392-020-00315-3. https://www.nature.com/articles/s41392-020-00315-3.
About Sartorius
Sartorius accelerates the development of breakthrough therapeutics with innovative solutions for lab filtration, cell/protein analysis, and more. Visit sartorius.com/biologics to ensure confidence in your biotherapeutic discovery and development program.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.net, which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.