SARS-CoV-2’s spike protein enables viral entry into the target cell through the exploitation of the Angiotensin-converting enzyme 2 (ACE2) receptor. The ACE2 gene is exceedingly conserved in mammalian animals at the DNA and peptide level, making them potential hosts for SARS-CoV-2.
A recent study discovered that the ACE2 gene was present in twelve common mammals. Amongst these mammals, humans were found to be the primary host of SARS-CoV-2. Mice are generally used as experimental animal models, with bats and pangolins suspected to be the intermediate host. Cats, ferrets, dogs and hamsters are commonly pets while pigs, cattle, goats and rabbits are generally found as livestock.1
As well as these twelve mammals, paguma larvata - a critical host of SARS - also possesses ACE2 expression.2
While ACE2 is expressed in mice, it is not present in any type of lung cells - only in the skin and tongue. It is therefore implied that the expression pattern of ACE2 protein in mammals could be species-specific.
Amino acid residues are found on the binding surface of ACE2, and these are necessary for virus recognition. Mutations within these core amino acid residues could lead to a difference in sensitivity for cross-species infection.
Seven key differences have been discovered between the sequence of human ACE2 and mouse or rat ACE2. These differences may be the reason that mouse or rat ACE2 protein does not bind to SARS-CoV-2 RBD protein. Because of this, rodent models like rats and mice may not be appropriate for SARS-CoV-2 research.
Studies have shown that the ACE2 gene is highly expressed on the cornea and skin dogs. This may result in dogs bring highly susceptible to SARS-CoV-2. Furthermore, as shown in the ACE2 phylogenetic tree below (Figure 1), dogs are genetically closer to humans than they are other mammals. This suggests that they are more reliable as an animal model for COVID-19 studies.1
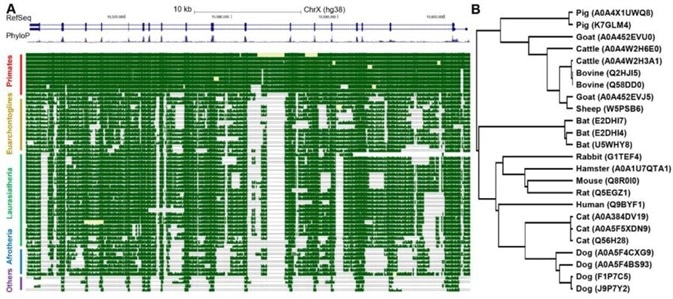
Figure 1. Conservation analysis of ACE2 gene. Image Credit: Sun et al., 2020 Atlas of ACE2 gene expression in mammals reveals novel insights in transmission of SARS-Cov-2
As well as dogs, apes and monkeys are also vulnerable to SARS-CoV-2. Numerous infection studies have demonstrated that rhesus monkeys can be successfully infected with SARS-CoV-2, causing them to develop COVID-19 like symptoms.
Cynomolgus ACE2 (Cat.No AC2-C52H7) created by ACRO has been proven to have good quality binding activity with the SARS-CoV-2 S1 protein and S RBD protein. This also confirms that cynomolgus ACE2 is extremely homologous with Human ACE2.3
Overall, dogs and monkeys are considered to be better animal models for COVID-19 research.
Paguma larvata is also a host of SARS. Its ACE2 receptor protein demonstrated an exceptional binding ability to the S protein of SARS-CoV-2, though this was not found to be comparable to human and cynomolgus ACE2. An ELISA assay was conducted in order to verify this result.
As shown in Figure 2 (below), the EC50s of paguma larvata ACE2 to SARS-CoV-2 S1 protein and S RBD protein were found to be 52 nM and 67 nM, respectively.
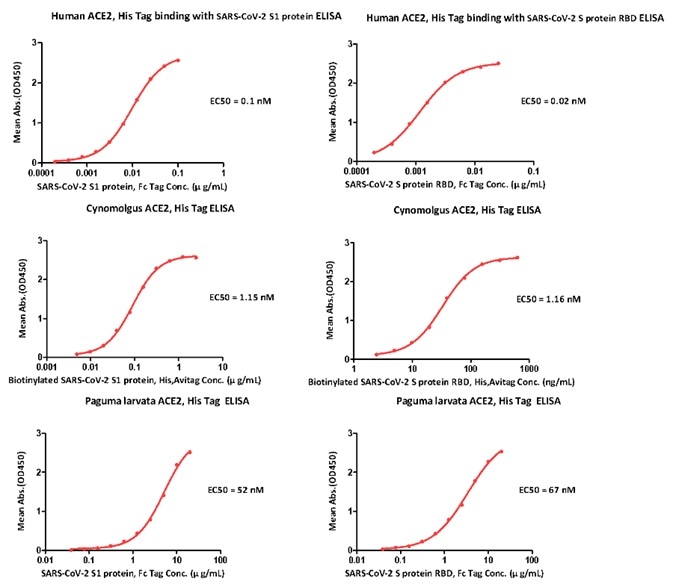
Figure 2. ELISA Binding activity results. Image Credit: ACROBiosystems
Meanwhile, the EC50s of cynomolgus ACE2 to SARS-CoV-2 S1 protein and S RBD protein were found to be 0.1 nM and 0.02 nM, respectively.
Finally, the EC50s of human ACE2 to SARS-CoV-2 S1 protein and S RBD protein were found to be 1.15 nM and 1.16 nM, respectively.
An investigation was undertaken into the binding activity between the ACE2 of human, cynomolgus or paguma larvata, alongside the S RBD of SARS-CoV-2 using BLI. Figure 3 outlines the results of this investigation. As can be seen, the binding activities of human and cynomolgus ACE2 to the S RBD of SARS-CoV2 are considerably higher than paguma larvata ACE2.
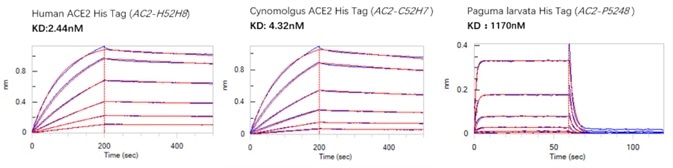
Figure 3. BLI binding activity results. Image Credit: ACROBiosystems
An SPR method was also utilized to examine the binding affinity between paguma larvata ACE2 and S RBD or S1 protein of SARS and SARS-CoV-2. Based on the results shown in Figure 4, it was determined that paguma larvata ACE2 is further susceptible to SARS.
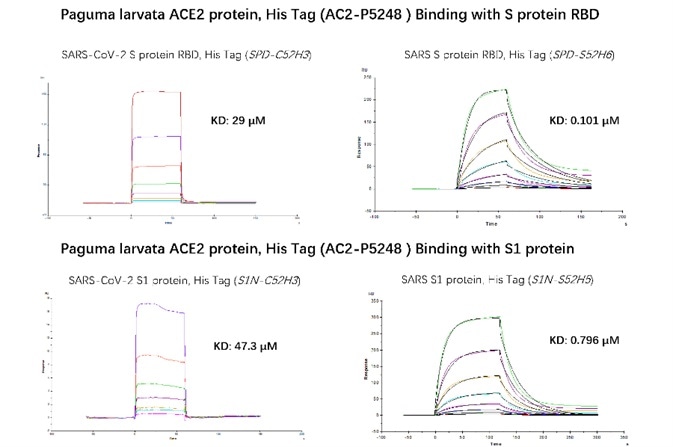
Figure 4. SPR binding activity results. Image Credit: ACROBiosystems
ACROBiosystems has developed effective ACE2 proteins for cynomolgus, mouse, rat, paguma larvata and humans. These proteins’ binding activity has been confirmed by BLI, meaning that these proteins can be employed in anti-SARS-CoV-2 research - see Figure 2.
Binding activity between the ACE2 of human, cynomolgus, paguma larvata and the S RBD of SARS-CoV-2 using BLI was also investigated. The results of this investigation are summarized in Figure 3. As can be seen here, the binding activities of human and cynomolgus ACE2 are far higher than those of paguma larvata.
References
- Sun et al., 2020 Atlas of ACE2 gene expression in mammals reveals novel insights in transmisson of SARS-Cov-2, doi: https://doi.org/10.1101/2020.03.30.015644
- Stawiski et al., 2020 Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. doi: https://doi.org/10.1101/2020.04.07.024752
- Melin et al., 2020 Comparative ACE2 variation and primate COVID-19 risk, https://doi.org/10.1101/2020.04.09.034967
- Wrapp et al., 2020 Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. DOI: 10.1126/science. Abb 2507
About ACROBiosystems
ACROBiosystems is a cornerstone enterprise of the pharmaceutical and biotechnology industries. Their mission is to help overcome challenges with innovative tools and solutions from discovery to the clinic. They supply life science tools designed to be used in discovery research and scalable to the clinical phase and beyond. By consistently adapting to new regulatory challenges and guidelines, ACROBiosystems delivers solutions, whether it comes through recombinant proteins, antibodies, assay kits, GMP-grade reagents, or custom services. ACROBiosystems empower scientists and engineers dedicated towards innovation to simplify and accelerate the development of new, better, and more affordable medicine.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.