The COVID-19 pandemic began over a year ago, and much research into SARS-CoV-2 has led to the development of therapeutics and vaccines. This article reviews some knowledge about SARS-CoV-2 and COVID-19, the developments in fighting the pandemic and the threat of emerging variants.
Targeting Spike protein for effective vaccine and therapeutics against SARS-CoV-2
Video Credit: ACROBiosystems
To date, COVID-19 has infected 100 million people and led to 2.2 million deaths worldwide, as seen in Figure 1A. SARS-CoV-2, the viral agent of this pandemic, is an enveloped virus with a single-strand positive-sense RNA genome.
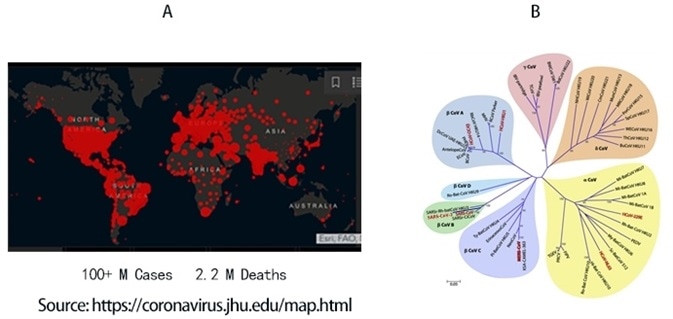
Figure 1. A) Global map of SARS-CoV-2 infection to date. B) Phylogenetic tree of coronaviruses. SARS-CoV-2 belongs to α-coronavirus clade in the B lineage. Image Credit: ACROBiosystems
Coronaviruses are a family of viruses that have a distinct morphology of crown-like projections infecting mammals and birds are divided into four genera: α, β, γ, and δ, as seen in Figure 1B.
Until the outbreak of the SARS epidemics of 2002, human coronaviruses would usually cause mild upper respiratory tract (UTR) infections. The SARS, followed by the MERS epidemic, demonstrated the ability of these coronaviruses to jump hosts and cause serious lower respiratory tract infection.
While SARS-CoV, SARS-CoV-2, and MERS-CoV are part of the β-coronavirus family, key differences in SARS-CoV-2 structural proteins have allowed it to explode in transmission rapidly and has gripped the world with a once-in-a-century pandemic, as seen in Table 1.
Table 1. Characteristics of the three recent coronavirus outbreaks2, 3. Source: ACROBiosystems
Fast research into the structural basis of SARS-CoV-2 host cell interaction and the mechanism of viral entry has only been viable due to previous SARS and MERS outbreak studies.
There are considerable differences between SARS-CoV and SARS-CoV-2, which explain the high transmission rates of SARS-CoV-21, even though both bind the same receptor on host cells via similar mechanisms.
Compared with coronavirus pandemics in the past, SARS-CoV-2 has been able to spread around the world quickly due to longer incubation and lower hospitalization rates, a novel furin cleavage site and no delay between symptom onset and maximum infectivity, as seen in Table 1.
Rapid advancement in understanding the structural underpinning of SARS-CoV-2 infectivity and transmissibility has shown promising therapeutics and vaccine candidates. Continual examination of viral mechanisms and the effects of emerging mutations are key to turning the tide on this pandemic.
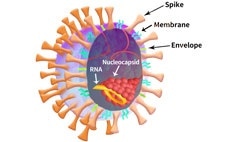
Figure 2. SARS-CoV-2 particle schematic. Image Credit: ACROBiosystems
As shown in Figure 2, The SARS-CoV-2 viral particle consists of a lipid of lipid bilayer containing envelop (E) protein, membrane (M) protein, nucleocapsid (N) protein, and the Spike (S) protein.
Of the structural proteins, viral fusion protein plays a key part in facilitating envelop viral particle entry and by proxy virus genetic material into the host cell.4 The S protein is the viral fusion glycoprotein trimer that binds to the host cell receptor and enables viral entry in SARS-CoV-2.
Host cell machinery is hijacked for replication, transcription, and assembly of viral particles upon entry, propagating its spread. When expressed in the host cell membrane, the viral fusion protein can lead to cell-cell fusion contributing to viral spread and virulence.4
The thermodynamic barrier of merging enveloped virus membrane to the host cell plasma membrane, in particular, is overcome by the kinetic energy stored in the fusion protein.4
In the majority of cases, the prefusion state of the fusion protein is metastable, and triggering events convert it to pre-hairpin intermediate, which exposes the hydrophobic fusion peptide embedding into the host cell membrane.4
Based on their structures and triggering mechanism, the fusion proteins can be divided into three classes. Class I consists primarily of α helices, while class II is mainly made up of β sheets, and class iii contains a mixture of α helices and β sheets4.
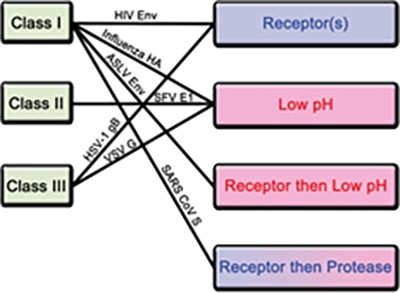
Figure 3. Classification of fusion proteins and triggering mechanisms with examples of corresponding viruses. Image Credit: ACROBiosystems
For fusion protein, there are four known triggering mechanisms island these include low pH, receptor binding, receptor binding followed by low pH, and receptor binding followed by protease cleavage, as seen in Figure 3.
Both class I and III fusion proteins are triggered by numerous mechanisms. The S protein of novel SARS-CoV-2 is a class I fusion protein triggered by receptor binding and protease cleavage.5
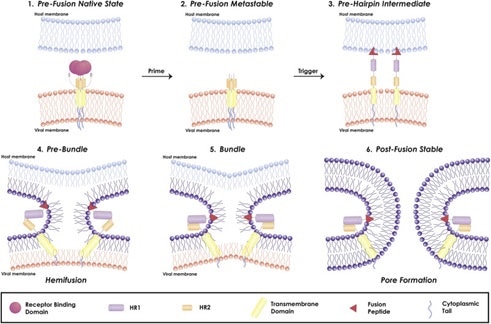
Figure 4. Mechanism of viral fusion entry primarily for class I fusion proteins. Step 4-6 applies to other classes of fusion proteins as well4. Image Credit: ACROBiosystems
The mechanism of a class I fusion protein starts with the pre-fusion state where the receptor binding subunit is clamped to the fusion subunit. The fusion protein can bind to a single or multiple receptor proteins.
Triggering of the fusion protein after receptor binding and protease cleaving allows the receptor-binding subunit to move out of the way to enable the fusion subunit to form a pre-hairpin with target membrane via fusion peptide, seen in Figure 4.
The pre-hairpin folds back because of this and causes N and C alpha helix heptad repeats to form a 6-helix bundling pulling the two membranes through close apposition, hemifusion, and fusion pore formation, as demonstrated in Figure 4.
Although enveloped viruses use many different varieties of fusion proteins and triggering mechanisms, all of them converge in these three last steps to gain entry into the cells. Examination of the viral fusion entry for SARS-CoV-2 is key to designing effective vaccines and developing effective therapies against COVID-19.
ACROBiosystems has developed a comprehensive line of critical reagents, including recombinant proteins, kits, antibodies, beads, etc., to cover the critical targets of SARS-CoV-2.
References
- B, H.; H, G.; P, Z.; ZL, S., Characteristics of SARS-CoV-2 and COVID-19. Nature reviews. Microbiology 2020.
- E, P.; M, K.; U, G.; DH, H.; N, P.; F, C.; M, S.; S, A. K.; L, S., Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. The Lancet. Infectious diseases 2020, 20 (9).
- Z, Z.; X, L.; X, S.; W, W.; GA, M.; Y, Z., From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respiratory research 2020, 21 (1).
- JM, W.; SE, D.; M, B.; K, S., Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Critical reviews in biochemistry and molecular biology 2008, 43 (3).
- T, T.; M, B.; JA, J.; GR, W.; S, D., Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral research 2020, 178.
About ACROBiosystems
ACROBiosystems is a cornerstone enterprise of the pharmaceutical and biotechnology industries. Their mission is to help overcome challenges with innovative tools and solutions from discovery to the clinic. They supply life science tools designed to be used in discovery research and scalable to the clinical phase and beyond. By consistently adapting to new regulatory challenges and guidelines, ACROBiosystems delivers solutions, whether it comes through recombinant proteins, antibodies, assay kits, GMP-grade reagents, or custom services. ACROBiosystems empower scientists and engineers dedicated towards innovation to simplify and accelerate the development of new, better, and more affordable medicine.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.