SARS-CoV-2 possesses a Spike protein which aids the viral entry into target cell exploiting Angiotensin-converting enzyme 2 (ACE2) receptor.[1][2] Located on the S1 subunit of SARS-CoV-2, the Spike protein receptor-binding domain (RBD) is the vital determinant of infectivity and viral tropism.
Under high positive selection pressure, the genome analysis of global SARS-CoV-2 strains has 32 RBD mutant strains clustering into 10 mutation types.
Three mutation types circulating in France, Wuhan, Hong Kong, and Shenzhen exhibited enhanced structural stability together with higher human ACE2 receptor affinity of the spike protein, suggesting that these mutants could have attained higher infectivity to humans.[3]
SARS-CoV-2 RBD mutation mapping
As it is the only domain to bind human ACE2 and initiate cell entry, it is thought that the RBD should be highly conserved. Polymorphism and divergence analysis revealed that, surprisingly, the RBD sequences were as diverse as the other regions of the S protein.
Of the 1609 SARS-CoV-2 strains in the public database, 32 strains possessed amino acid mutations in the RBD. These mutant strains were reported from a number of locations. These include Belgium, Finland, France, China, the UK, the USA, and India, the majority of which possessed just one amino acid change compared to the originally reported genome.
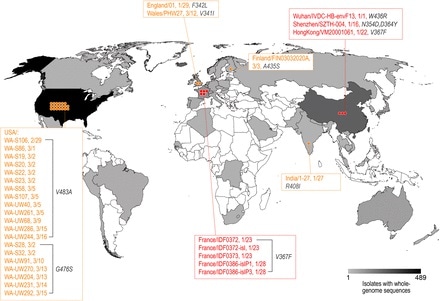
Figure 1. Distribution of the SARS-CoV-2 strains mutated in the RBD of the S protein. The geographic distribution of the RBD mutant strains is displayed. The strains with names highlighted in red are mutants with the enhanced binding affinity. The strains with names noted in yellow are mutants with similar binding affinities. Image Credit: bioRxiv preprint doi: https://doi.org/10.1101/2020.03.15.991844
Three mutation types bind human ACE2 receptor with higher affinity
In order to estimate the functional change suggested by the RBD mutations molecular dynamics simulations were carried out. Three RBD mutant types (N354D and D364Y, V367F, W436R) showed much lower binding free energy (ΔG), indicating a significantly higher affinity to human ACE2.[3]
Structural basis for increased affinity
The binding surface of the RBD to ACE2 lacks structural rigidity as it is mostly in random coil conformation. So, a firm scaffold should help to maintain this conformation of the interaction surface, and could facilitate the binding affinity.
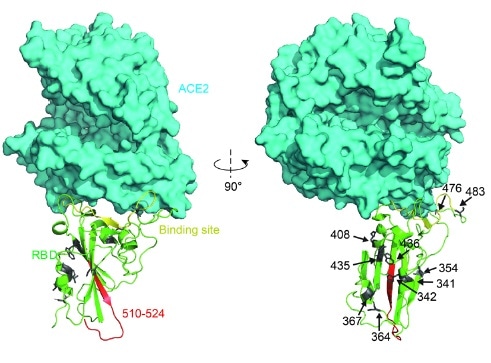
Figure 2. Structural analysis of RBD mutants and the effects on their binding affinity. Spatial 409 location of the mutant amino acids and the fragment 510-524. Image Credit: bioRxiv preprint doi: https://doi.org/10.1101/2020.03.15.991844
Centered by residues 510-524 (Fig. 2, marked as red), the beta-sheet structure scaffold supplies this rigidity. “Higher affinity” mutants (N354D D364Y, V367F, and W436R) generally showed a lower ΔG in the binding site region, which heightened the structural stability.
Furthermore, the mutation W436R supplied a positively charged Arg in the proximity of the complementing highly negative charged ACE2 surface. This potential electrostatic attraction may contribute to the higher affinity.[3]
To investigate the findings discussed above, ACROBiosystems expressed the recombinant prototype spike protein RBD and “higher affinity” mutants in house. They used three different methods, including ELISA, Biacore SPR, and Octet BLI, to analyze the binding affinities between human ACE2 protein and different spike proteins respectively. The results were summarized in Fig. 3, Fig. 4, and Fig. 5 below. As you can see, all the “higher affinity” spike protein RBD mutants showed stronger binding affinities compared to the prototype.
Bioactivity-ELISA
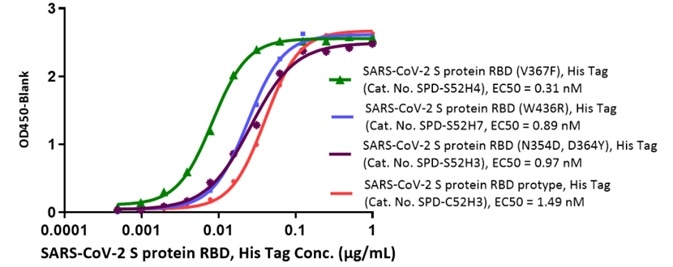
Figure 3. Bioactivity- ELISA results. Human ACE2, Fc Tag (Cat. No. AC2-H5257) was immobilized on the plate. RBD (V367F) EC50: 0.31 nM > RBD (W436R) EC50: 0.89 nM > RBD (N354D, D364Y) EC50: 0.97 nM > prototype RBD EC50: 1.47 nM. Image Credit: ACROBiosystems
Bioactivity-BLI
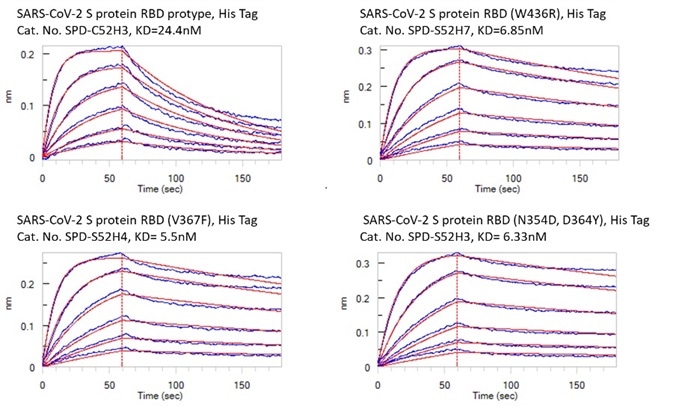
Figure 4. Bioactivity – Octet BLI results. The KD between Human ACE2 and prototype spike protein RBD is 24.4 nM. The KD between Human ACE2 and RBD(V367F), RBD (W436R), and RBD (N354D, D364Y) were 5.5 nM, 6.85 nM, and 6.33 nM respectively. Image Credit: ACROBiosystems
Bioactivity-SPR
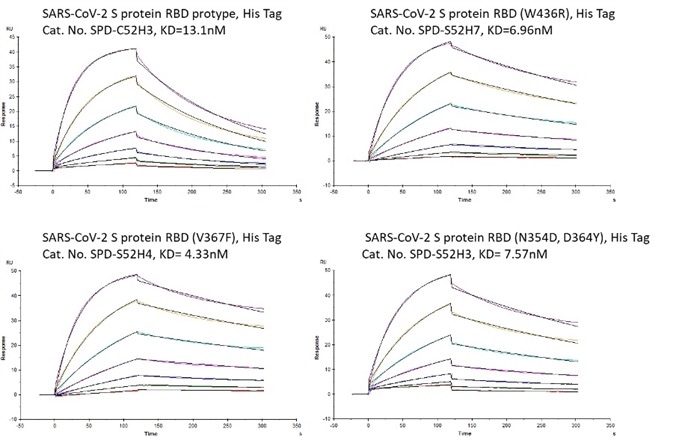
Figure 5. Bioactivity – Biacore SPR results. The KD between Human ACE2 and prototype spike protein RBD is 13.1 nM. The KD between Human ACE2 and RBD(V367F), RBD (W436R), and RBD (N354D, D364Y) were 4.33 nM, 6.96 nM, and 7.57 nM respectively. Image Credit: ACROBiosystems
References
- Hoffmann et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, 2020, Cell 181, 271–280;
- Zhou et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, 2020, Nature 579, 270-273;
- Ou et al. Emergence of RBD mutations in circulating SARS-CoV-2 strains enhancing the structural stability and human ACE2 receptor affinity of the spike protein, bioRxiv preprint doi: https://doi.org/10.1101/2020.03.15.991844;
About ACROBiosystems
ACROBiosystems is a cornerstone enterprise of the pharmaceutical and biotechnology industries. Their mission is to help overcome challenges with innovative tools and solutions from discovery to the clinic. They supply life science tools designed to be used in discovery research and scalable to the clinical phase and beyond. By consistently adapting to new regulatory challenges and guidelines, ACROBiosystems delivers solutions, whether it comes through recombinant proteins, antibodies, assay kits, GMP-grade reagents, or custom services. ACROBiosystems empower scientists and engineers dedicated towards innovation to simplify and accelerate the development of new, better, and more affordable medicine.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.